Inferring circadian gene regulatory relationships from gene expression data with a hybrid framework
- PMID: 37752445
- PMCID: PMC10521455
- DOI: 10.1186/s12859-023-05458-y
Inferring circadian gene regulatory relationships from gene expression data with a hybrid framework
Abstract
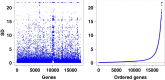
Background: The central biological clock governs numerous facets of mammalian physiology, including sleep, metabolism, and immune system regulation. Understanding gene regulatory relationships is crucial for unravelling the mechanisms that underlie various cellular biological processes. While it is possible to infer circadian gene regulatory relationships from time-series gene expression data, relying solely on correlation-based inference may not provide sufficient information about causation. Moreover, gene expression data often have high dimensions but a limited number of observations, posing challenges in their analysis.
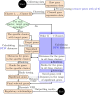
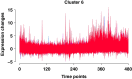
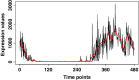
Methods: In this paper, we introduce a new hybrid framework, referred to as Circadian Gene Regulatory Framework (CGRF), to infer circadian gene regulatory relationships from gene expression data of rats. The framework addresses the challenges of high-dimensional data by combining the fuzzy C-means clustering algorithm with dynamic time warping distance. Through this approach, we efficiently identify the clusters of genes related to the target gene. To determine the significance of genes within a specific cluster, we employ the Wilcoxon signed-rank test. Subsequently, we use a dynamic vector autoregressive method to analyze the selected significant gene expression profiles and reveal directed causal regulatory relationships based on partial correlation.
Conclusion: The proposed CGRF framework offers a comprehensive and efficient solution for understanding circadian gene regulation. Circadian gene regulatory relationships are inferred from the gene expression data of rats based on the Aanat target gene. The results show that genes Pde10a, Atp7b, Prok2, Per1, Rhobtb3 and Dclk1 stand out, which have been known to be essential for the regulation of circadian activity. The potential relationships between genes Tspan15, Eprs, Eml5 and Fsbp with a circadian rhythm need further experimental research.
Keywords: Circadian gene; Dynamic time warping; Fuzzy c-means clustering; Gene expression data; Gene regulatory relationships.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Network motif-based identification of transcription factor-target gene relationships by integrating multi-source biological data.BMC Bioinformatics. 2008 Apr 21;9:203. doi: 10.1186/1471-2105-9-203. BMC Bioinformatics. 2008. PMID: 18426580 Free PMC article.
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
A network biology study on circadian rhythm by integrating various omics data.OMICS. 2009 Aug;13(4):313-24. doi: 10.1089/omi.2009.0040. OMICS. 2009. PMID: 19645592
-
Melatonin synthesis and clock gene regulation in the pineal organ of teleost fish compared to mammals: Similarities and differences.Gen Comp Endocrinol. 2019 Aug 1;279:27-34. doi: 10.1016/j.ygcen.2018.07.010. Epub 2018 Jul 17. Gen Comp Endocrinol. 2019. PMID: 30026020 Review.
-
Invited review: regulation of mammalian circadian clock genes.J Appl Physiol (1985). 2002 Mar;92(3):1348-55. doi: 10.1152/japplphysiol.00759.2001. J Appl Physiol (1985). 2002. PMID: 11842077 Review.
Cited by
-
GRLGRN: graph representation-based learning to infer gene regulatory networks from single-cell RNA-seq data.BMC Bioinformatics. 2025 Apr 18;26(1):108. doi: 10.1186/s12859-025-06116-1. BMC Bioinformatics. 2025. PMID: 40251476 Free PMC article.
-
Integrative analysis of ASXL family genes reveals ASXL2 as an immunoregulatory molecule in head and neck squamous cell carcinoma.Sci Rep. 2024 Dec 28;14(1):31368. doi: 10.1038/s41598-024-82815-8. Sci Rep. 2024. PMID: 39732849 Free PMC article.
References
-
- Steuer R. On the analysis and interpretation of correlations in metabolomic data. Brief Bioinform. 2006;7(2):151–158. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Statist Soc B. 1995;57:289–300.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

