Machine learning-derived identification of tumor-infiltrating immune cell-related signature for improving prognosis and immunotherapy responses in patients with skin cutaneous melanoma
- PMID: 37752452
- PMCID: PMC10521465
- DOI: 10.1186/s12935-023-03048-9
Machine learning-derived identification of tumor-infiltrating immune cell-related signature for improving prognosis and immunotherapy responses in patients with skin cutaneous melanoma
Abstract
Background: Immunoblockade therapy based on the PD-1 checkpoint has greatly improved the survival rate of patients with skin cutaneous melanoma (SKCM). However, existing anti-PD-1 therapeutic efficacy prediction markers often exhibit a poor situation of poor reliability in identifying potential beneficiary patients in clinical applications, and an ideal biomarker for precision medicine is urgently needed.
Methods: 10 multicenter cohorts including 4 SKCM cohorts and 6 immunotherapy cohorts were selected. Through the analysis of WGCNA, survival analysis, consensus clustering, we screened 36 prognostic genes. Then, ten machine learning algorithms were used to construct a machine learning-derived immune signature (MLDIS). Finally, the independent data sets (GSE22153, GSE54467, GSE59455, and in-house cohort) were used as the verification set, and the ROC index standard was used to evaluate the model.
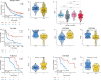
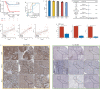
Results: Based on computing framework, we found that patients with high MLDIS had poor overall survival and has good prediction performance in all cohorts and in-house cohort. It is worth noting that MLDIS performs better in each data set than almost all models which from 51 prognostic signatures for SKCM. Meanwhile, high MLDIS have a positive prognostic impact on patients treated with anti-PD-1 immunotherapy by driving changes in the level of infiltration of immune cells in the tumor microenvironment. Additionally, patients suffering from SKCM with high MLDIS were more sensitive to immunotherapy.
Conclusions: Our study identified that MLDIS could provide new insights into the prognosis of SKCM and predict the immunotherapy response in patients with SKCM.
Keywords: Immunotherapy; Machine learning; Skin cutaneous melanoma; Tumor microenvironment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Garbe C, Amaral T, Peris K, Hauschild A, Arenberger P, Basset-Seguin N, Bastholt L, Bataille V, Del Marmol V, Dréno B, et al. European consensus-based interdisciplinary guideline for melanoma. Part 2: treatment—update 2022. Eur J Cancer (Oxford, England: 1990) 2022;170:256–284. doi: 10.1016/j.ejca.2022.04.018. - DOI - PubMed
LinkOut - more resources
Full Text Sources

