Type I/type III IFN and related factors regulate JEV infection and BBB endothelial integrity
- PMID: 37752509
- PMCID: PMC10523659
- DOI: 10.1186/s12974-023-02891-x
Type I/type III IFN and related factors regulate JEV infection and BBB endothelial integrity
Abstract
Background: Japanese encephalitis virus (JEV) remains a predominant cause of Japanese encephalitis (JE) globally. Its infection is usually accompanied by disrupted blood‒brain barrier (BBB) integrity and central nervous system (CNS) inflammation in a poorly understood pathogenesis. Productive JEV infection in brain microvascular endothelial cells (BMECs) is considered the initial event of the virus in penetrating the BBB. Type I/III IFN and related factors have been described as negative regulators in CNS inflammation, whereas their role in JE remains ambiguous.
Methods: RNA-sequencing profiling (RNA-seq), real-time quantitative PCR, enzyme-linked immunosorbent assay, and Western blotting analysis were performed to analyze the gene and protein expression changes between mock- and JEV-infected hBMECs. Bioinformatic tools were used to cluster altered signaling pathway members during JEV infection. The shRNA-mediated immune factor-knockdown hBMECs and the in vitro transwell BBB model were utilized to explore the interrelation between immune factors, as well as between immune factors and BBB endothelial integrity.
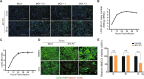
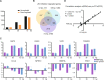
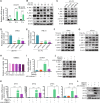
Results: RNA-Seq data of JEV-infected hBMECs identified 417, 1256, and 2748 differentially expressed genes (DEGs) at 12, 36, and 72 h post-infection (hpi), respectively. The altered genes clustered into distinct pathways in gene ontology (GO) terms and KEGG pathway enrichment analysis, including host antiviral immune defense and endothelial cell leakage. Further investigation revealed that pattern-recognition receptors (PRRs, including TLR3, RIG-I, and MDA5) sensed JEV and initiated IRF/IFN signaling. IFNs triggered the expression of interferon-induced proteins with tetratricopeptide repeats (IFITs) via the JAK/STAT pathway. Distinct PRRs exert different functions in barrier homeostasis, while treatment with IFN (IFN-β and IFN-λ1) in hBMECs stabilizes the endothelial barrier by alleviating exogenous destruction. Despite the complex interrelationship, IFITs are considered nonessential in the IFN-mediated maintenance of hBMEC barrier integrity.
Conclusions: This research provided the first comprehensive description of the molecular mechanisms of host‒pathogen interplay in hBMECs responding to JEV invasion, in which type I/III IFN and related factors strongly correlated with regulating the hBMEC barrier and restricting JEV infection. This might help with developing an attractive therapeutic strategy in JE.
Keywords: Human brain microvascular endothelial cells; IFITs; IFNs; Japanese encephalitis virus; PRRs; RNA-Seq.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
Japanese encephalitis virus NS1 and NS1' protein disrupts the blood-brain barrier through macrophage migration inhibitory factor-mediated autophagy.J Virol. 2024 May 14;98(5):e0011624. doi: 10.1128/jvi.00116-24. Epub 2024 Apr 9. J Virol. 2024. PMID: 38591880 Free PMC article.
-
Viral Infection of the Central Nervous System and Neuroinflammation Precede Blood-Brain Barrier Disruption during Japanese Encephalitis Virus Infection.J Virol. 2015 May;89(10):5602-14. doi: 10.1128/JVI.00143-15. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762733 Free PMC article.
-
Palmitoylation of hIFITM1 inhibits JEV infection and contributes to BBB stabilization.Int J Biol Macromol. 2024 Mar;262(Pt 1):129731. doi: 10.1016/j.ijbiomac.2024.129731. Epub 2024 Jan 24. Int J Biol Macromol. 2024. PMID: 38278394
-
Pathogenicity and virulence of Japanese encephalitis virus: Neuroinflammation and neuronal cell damage.Virulence. 2021 Dec;12(1):968-980. doi: 10.1080/21505594.2021.1899674. Virulence. 2021. PMID: 33724154 Free PMC article. Review.
-
Molecular Mechanism and Role of Japanese Encephalitis Virus Infection in Central Nervous System-Mediated Diseases.Viruses. 2022 Nov 30;14(12):2686. doi: 10.3390/v14122686. Viruses. 2022. PMID: 36560690 Free PMC article. Review.
Cited by
-
Common Chemical Plasticizer Di(2-Ethhylhexyl) Phthalate Exposure Exacerbates Coxsackievirus B3 Infection.Viruses. 2024 Nov 23;16(12):1821. doi: 10.3390/v16121821. Viruses. 2024. PMID: 39772131 Free PMC article.
-
Japanese encephalitis virus NS1 and NS1' protein disrupts the blood-brain barrier through macrophage migration inhibitory factor-mediated autophagy.J Virol. 2024 May 14;98(5):e0011624. doi: 10.1128/jvi.00116-24. Epub 2024 Apr 9. J Virol. 2024. PMID: 38591880 Free PMC article.
-
Multiomics Analysis Reveals Role of ncRNA in Hypoxia of Mouse Brain Microvascular Endothelial Cells.Int J Mol Sci. 2025 Jun 12;26(12):5629. doi: 10.3390/ijms26125629. Int J Mol Sci. 2025. PMID: 40565093 Free PMC article.
-
Japanese encephalitis virus NS1 inhibits IFN-β production by interacting with DDX3X.J Virol. 2025 May 20;99(5):e0007725. doi: 10.1128/jvi.00077-25. Epub 2025 Apr 15. J Virol. 2025. PMID: 40231822 Free PMC article.
References
-
- Kaur R, Vrati S. Development of a recombinant vaccine against Japanese encephalitis. J Neurovirol. 2003;9:421–431. - PubMed
-
- Eigenmann DE, Xue G, Kim KS, Moses AV, Hamburger M, Oufir M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studies. Fluids Barriers CNS. 2013;10:33. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

