Structural biology of complement receptors
- PMID: 37753090
- PMCID: PMC10518620
- DOI: 10.3389/fimmu.2023.1239146
Structural biology of complement receptors
Abstract
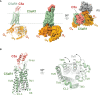
The complement system plays crucial roles in a wide breadth of immune and inflammatory processes and is frequently cited as an etiological or aggravating factor in many human diseases, from asthma to cancer. Complement receptors encompass at least eight proteins from four structural classes, orchestrating complement-mediated humoral and cellular effector responses and coordinating the complex cross-talk between innate and adaptive immunity. The progressive increase in understanding of the structural features of the main complement factors, activated proteolytic fragments, and their assemblies have spurred a renewed interest in deciphering their receptor complexes. In this review, we describe what is currently known about the structural biology of the complement receptors and their complexes with natural agonists and pharmacological antagonists. We highlight the fundamental concepts and the gray areas where issues and problems have been identified, including current research gaps. We seek to offer guidance into the structural biology of the complement system as structural information underlies fundamental and therapeutic research endeavors. Finally, we also indicate what we believe are potential developments in the field.
Keywords: C5aR1/C5L2/C3aR; CR1/CR2; CR3/CR4; CRIg; complement; complement receptors; host-pathogen interactions; structural biology.
Copyright © 2023 Santos-López, de la Paz, Fernández and Vega.
Conflict of interest statement
Abvance Biotech SL provided salaries for KdlP and FJF. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



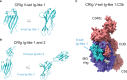
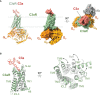
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

