Group-level brain decoding with deep learning
- PMID: 37753636
- PMCID: PMC10619368
- DOI: 10.1002/hbm.26500
Group-level brain decoding with deep learning
Abstract
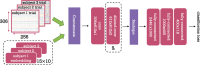
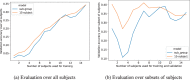
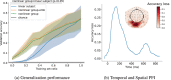
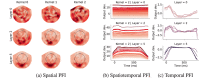
Decoding brain imaging data are gaining popularity, with applications in brain-computer interfaces and the study of neural representations. Decoding is typically subject-specific and does not generalise well over subjects, due to high amounts of between subject variability. Techniques that overcome this will not only provide richer neuroscientific insights but also make it possible for group-level models to outperform subject-specific models. Here, we propose a method that uses subject embedding, analogous to word embedding in natural language processing, to learn and exploit the structure in between-subject variability as part of a decoding model, our adaptation of the WaveNet architecture for classification. We apply this to magnetoencephalography data, where 15 subjects viewed 118 different images, with 30 examples per image; to classify images using the entire 1 s window following image presentation. We show that the combination of deep learning and subject embedding is crucial to closing the performance gap between subject- and group-level decoding models. Importantly, group models outperform subject models on low-accuracy subjects (although slightly impair high-accuracy subjects) and can be helpful for initialising subject models. While we have not generally found group-level models to perform better than subject-level models, the performance of group modelling is expected to be even higher with bigger datasets. In order to provide physiological interpretation at the group level, we make use of permutation feature importance. This provides insights into the spatiotemporal and spectral information encoded in the models. All code is available on GitHub (https://github.com/ricsinaruto/MEG-group-decode).
Keywords: MEG; decoding; deep learning; neuroimaging; permutation feature importance; transfer learning.
© 2023 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
The authors report no conflict of interest.
Figures



References
-
- Altmann, A. , Toloşi, L. , Sander, O. , & Lengauer, T. (2010). Permutation importance: A corrected feature importance measure. Bioinformatics, 26(10), 1340–1347. - PubMed
-
- Benz, K. R. (2020). Hyperalignment in meg: A first implementation using auditory evoked fields.
-
- Borovykh, A. , Bohte, S. , & Oosterlee, C. W. (2018). Dilated convolutional neural networks for time series forecasting. Journal of Computational Finance, 22(4), 73–101.
-
- Brown, T. , Mann, B. , Ryder, N. , Subbiah, M. , Kaplan, J. D. , Dhariwal, P. , Neelakantan, A. , Shyam, P. , Sastry, G. , Askell, A. , Agarwal, S. , Herbert‐Voss, A. , Krueger, G. , Henighan, T. , Child, R. , Ramesh, A. , Ziegler, D. , Wu, J. , Winter, C. , … Amodei, D. (2020). Language models are few‐shot learners. In Larochelle H., Ranzato M., Hadsell R., Balcan M., & Lin H. (Eds.), Advances in Neural Information Processing Systems (Vol. 33, pp. 1877–1901). Curran Associates, Inc.
-
- Chehab, O. , Defossez, A. , Loiseau, J.‐C. , Gramfort, A. , & King, J.‐R. (2022). Deep recurrent encoder: A scalable end‐to‐end network to model brain signals (p. 1). Neurons.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

