Toward Continuous Molecular Testing Using Gold-Coated Threads as Multi-Target Electrochemical Biosensors
- PMID: 37754078
- PMCID: PMC10526339
- DOI: 10.3390/bios13090844
Toward Continuous Molecular Testing Using Gold-Coated Threads as Multi-Target Electrochemical Biosensors
Abstract
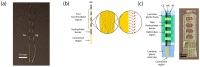
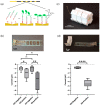
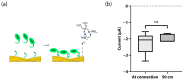
Analytical systems based on isothermal nucleic acid amplification tests (NAATs) paired with electroanalytical detection enable cost-effective, sensitive, and specific digital pathogen detection for various in situ applications such as point-of-care medical diagnostics, food safety monitoring, and environmental surveillance. Self-assembled monolayers (SAMs) on gold surfaces are reliable platforms for electroanalytical DNA biosensors. However, the lack of automation and scalability often limits traditional chip-based systems. To address these challenges, we propose a continuous thread-based device that enables multiple electrochemical readings on a functionalized working electrode Au thread with a single connection point. We demonstrate the possibility of rolling the thread on a spool, which enables easy manipulation in a roll-to-roll architecture for high-throughput applications. As a proof of concept, we have demonstrated the detection of recombinase polymerase amplification (RPA) isothermally amplified DNA from the two toxic microalgae species Ostreopsis cf. ovata and Ostreopsis cf. siamensis by performing a sandwich hybridization assay (SHA) with electrochemical readout.
Keywords: chronoamperometry; isothermal DNA amplification; metal-coated threads; roll-to-roll; sandwich hybridization assay; self-assembled monolayers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Oliveira B.B., Veigas B., Baptista P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021;2:752600. doi: 10.3389/fsens.2021.752600. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

