Recent Progress in Single-Nucleotide Polymorphism Biosensors
- PMID: 37754098
- PMCID: PMC10527258
- DOI: 10.3390/bios13090864
Recent Progress in Single-Nucleotide Polymorphism Biosensors
Abstract
Single-nucleotide polymorphisms (SNPs), the most common form of genetic variation in the human genome, are the main cause of individual differences. Furthermore, such attractive genetic markers are emerging as important hallmarks in clinical diagnosis and treatment. A variety of destructive abnormalities, such as malignancy, cardiovascular disease, inherited metabolic disease, and autoimmune disease, are associated with single-nucleotide variants. Therefore, identification of SNPs is necessary for better understanding of the gene function and health of an individual. SNP detection with simple preparation and operational procedures, high affinity and specificity, and cost-effectiveness have been the key challenge for years. Although biosensing methods offer high specificity and sensitivity, as well, they suffer drawbacks, such as complicated designs, complicated optimization procedures, and the use of complicated chemistry designs and expensive reagents, as well as toxic chemical compounds, for signal detection and amplifications. This review aims to provide an overview on improvements for SNP biosensing based on fluorescent and electrochemical methods. Very recently, novel designs in each category have been presented in detail. Furthermore, detection limitations, advantages and disadvantages, and challenges have also been presented for each type.
Keywords: SNPs; biosensor; electrochemistry; fluorescence; quartz crystal microbalance (QCM).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
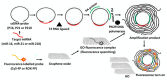
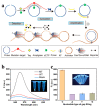
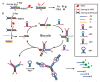
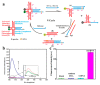
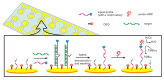


References
-
- Xu M., Wang X., Tian J., Chen J., Wei X., Li W. A clamp-improved universal amplified system for ratiometric fluorescent detection of single-nucleotide polymorphisms coupled with a novel dual-emissive silver nanocluster. Sens. Actuators B Chem. 2022;367:132151. doi: 10.1016/j.snb.2022.132151. - DOI
-
- Akekawatchai C., Phuegsilp C., Changsri K., Soimanee T., Sretapunya W. Genotypic and allelic distribution of IFN-gamma +874T/A and TGF-beta1 -509C/T single-nucleotide polymorphisms in human immunodeficiency virus-infected Thais. J. Med. Virol. 2022;94:2882–2886. doi: 10.1002/jmv.27567. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

