Acquisition of Type I methyltransferase via horizontal gene transfer increases the drug resistance of Aeromonas veronii
- PMID: 37754275
- PMCID: PMC10569733
- DOI: 10.1099/mgen.0.001107
Acquisition of Type I methyltransferase via horizontal gene transfer increases the drug resistance of Aeromonas veronii
Abstract
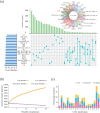
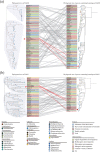
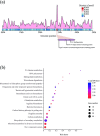
Aeromonas veronii is an opportunistic pathogen that affects both fish and mammals, including humans, leading to bacteraemia, sepsis, meningitis and even death. The increasing virulence and drug resistance of A. veronii are of significant concern and pose a severe risk to public safety. The Type I restriction-modification (RM) system, which functions as a bacterial defence mechanism, can influence gene expression through DNA methylation. However, little research has been conducted to explore its origin, evolutionary path, and relationship to virulence and drug resistance in A. veronii. In this study, we analysed the pan-genome of 233 A. veronii strains, and the results indicated that it was 'open', meaning that A. veronii has acquired additional genes from other species. This suggested that A. veronii had the potential to adapt and evolve rapidly, which might have contributed to its drug resistance. One Type I methyltransferase (MTase) and two complete Type I RM systems were identified, namely AveC4I, AveC4II and AveC4III in A. veronii strain C4, respectively. Notably, AveC4I was exclusive to A. veronii C4. Phylogenetic analysis revealed that AveC4I was derived from horizontal gene transfer from Thiocystis violascens and exchanged genes with the human pathogen Comamonas kerstersii. Single molecule real-time sequencing was applied to identify the motif methylated by AveC4I, which was unique and not recognized by any reported MTases in the REBASE database. We also annotated the functions and pathways of the genes containing the motif, revealing that AveC4I may control drug resistance in A. veronii C4. Our findings provide new insight on the mechanisms underlying drug resistance in pathogenic bacteria. By identifying the specific genes and pathways affected by AveC4I, this study may aid in the development of new therapeutic approaches to combat A. veronii infections.
Keywords: Aeromonas veronii; Type I restriction-modification system; drug resistance; horizontal gene transfer; methylome.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila.Antonie Van Leeuwenhoek. 2016 Jul;109(7):945-56. doi: 10.1007/s10482-016-0693-6. Epub 2016 Apr 13. Antonie Van Leeuwenhoek. 2016. PMID: 27075453
-
Complete genome sequence and genome-wide transposon mutagenesis enable the determination of genes required for sodium hypochlorite tolerance and drug resistance in pathogen Aeromonas veronii GD2019.Microbiol Res. 2024 Jul;284:127731. doi: 10.1016/j.micres.2024.127731. Epub 2024 Apr 17. Microbiol Res. 2024. PMID: 38653011
-
Characterization of Virulence Properties of Aeromonas veronii Isolated from Diseased Gibel Carp (Carassius gibelio).Int J Mol Sci. 2016 Apr 1;17(4):496. doi: 10.3390/ijms17040496. Int J Mol Sci. 2016. PMID: 27043558 Free PMC article.
-
Genome sequencing and annotation of multi-virulent Aeromonas veronii XhG1.2 isolated from diseased Xiphophorus hellerii.Genomics. 2021 Jan;113(1 Pt 2):991-998. doi: 10.1016/j.ygeno.2020.10.034. Epub 2020 Nov 2. Genomics. 2021. PMID: 33144215
-
Characterization of virulence properties and multi-drug resistance profiles in motile Aeromonas spp. isolated from zebrafish (Danio rerio).Lett Appl Microbiol. 2018 Dec;67(6):598-605. doi: 10.1111/lam.13075. Epub 2018 Nov 12. Lett Appl Microbiol. 2018. PMID: 30229985
Cited by
-
The complete genome sequence of unculturable Mycoplasma faucium obtained through clinical metagenomic next-generation sequencing.Front Cell Infect Microbiol. 2024 Apr 16;14:1368923. doi: 10.3389/fcimb.2024.1368923. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38694516 Free PMC article.
-
Genome-wide DNA N6-methyladenosine in Aeromonas veronii and Helicobacter pylori.BMC Genomics. 2024 Feb 8;25(1):161. doi: 10.1186/s12864-024-10074-y. BMC Genomics. 2024. PMID: 38331763 Free PMC article.
-
Targeted insights into Aeromonas hydrophila biofilms: Surface preferences, resistance mechanisms, and gene expression.Poult Sci. 2025 Apr;104(4):104851. doi: 10.1016/j.psj.2025.104851. Epub 2025 Jan 25. Poult Sci. 2025. PMID: 40043669 Free PMC article.
-
The emerging role of DNA methylation in the pathogenicity of bacterial pathogens.J Bacteriol. 2025 Aug 21;207(8):e0010825. doi: 10.1128/jb.00108-25. Epub 2025 Jul 17. J Bacteriol. 2025. PMID: 40673666 Free PMC article. Review.
-
The restriction impacts of the Type III restriction-modification system on the transmission dynamics of antimicrobial resistance genes in Campylobacter jejuni.Front Microbiol. 2025 Jul 17;16:1496275. doi: 10.3389/fmicb.2025.1496275. eCollection 2025. Front Microbiol. 2025. PMID: 40746326 Free PMC article.
References
-
- Roberts GA, Chen K, Cooper LP, White JH, Blakely GW, et al. Removal of a frameshift between the hsdM and hsdS genes of the EcoKI Type IA DNA restriction and modification system produces a new type of system and links the different families of type I systems. Nucleic Acids Res. 2012;40:10916–10924. doi: 10.1093/nar/gks876. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

