Rhizosphere and detritusphere habitats modulate expression of soil N-cycling genes during plant development
- PMID: 37754554
- PMCID: PMC10654102
- DOI: 10.1128/msystems.00315-23
Rhizosphere and detritusphere habitats modulate expression of soil N-cycling genes during plant development
Abstract
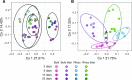
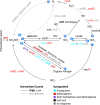
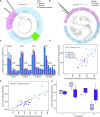
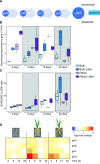
Plant roots modulate microbial nitrogen (N) cycling by regulating the supply of root-derived carbon and nitrogen uptake. These differences in resource availability cause distinct micro-habitats to develop: soil near living roots, decaying roots, near both, or outside the direct influence of roots. While many environmental factors and genes control the microbial processes involved in the nitrogen cycle, most research has focused on single genes and pathways, neglecting the interactive effects these pathways have on each other. The processes controlled by these pathways determine consumption and production of N by soil microorganisms. We followed the expression of N-cycling genes in four soil microhabitats over a period of active root growth for an annual grass. We found that the presence of root litter and living roots significantly altered gene expression involved in multiple nitrogen pathways, as well as tradeoffs between pathways, which ultimately regulate N availability to plants.
Keywords: detritusphere; gene expression; metatranscriptomics; nitrification; plant litter; rhizosphere; soil microbiome; soil nitrogen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Sokol NW, Slessarev E, Marschmann GL, Nicolas A, Blazewicz SJ, Brodie EL, Firestone MK, Foley MM, Hestrin R, Hungate BA, Koch BJ, Stone BW, Sullivan MB, Zablocki O, LLNL Soil Microbiome Consortium, Pett-Ridge J. 2022. Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat Rev Microbiol 20:415–430. doi: 10.1038/s41579-022-00695-z - DOI - PubMed
-
- Clarholm M. 1985. Interactions of bacteria, protozoa and plants leading to mineralization of soil nitrogen. Soil Biol Biochem 17:181–187. doi: 10.1016/0038-0717(85)90113-0 - DOI
-
- Koller R, Scheu S, Bonkowski M, Robin C. 2013. Protozoa stimulate N uptake and growth of arbuscular mycorrhizal plants. Soil Biol Biochem 65:204–210. doi: 10.1016/j.soilbio.2013.05.020 - DOI

