Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota)
- PMID: 37755002
- PMCID: PMC10532482
- DOI: 10.3390/jof9090894
Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota)
Abstract
Background: The delimitation of species of Tulasnella has been extensively studied, mainly at the morphological (sexual and asexual states) and molecular levels-showing ambiguity between them. An integrative species concept that includes characteristics such as molecular, ecology, morphology, and other information is crucial for species delimitation in complex groups such as Tulasnella.
Objectives: The aim of this study is to test evolutionary relationships using a combination of alignment-based and alignment-free distance matrices as an alternative molecular tool to traditional methods, and to consider the secondary structures and CBCs from ITS2 (internal transcribed spacer) sequences for species delimitation in Tulasnella.
Methodology: Three phylogenetic approaches were plotted: (i) alignment-based, (ii) alignment-free, and (iii) a combination of both distance matrices using the DISTATIS and pvclust libraries from an R package. Finally, the secondary structure consensus was modeled by Mfold, and a CBC analysis was obtained to complement the species delimitation using 4Sale.
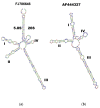
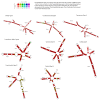
Results and conclusions: The phylogenetic tree results showed delimited monophyletic clades in Tulasnella spp., where all 142 Tulasnella sequences were divided into two main clades A and B and assigned to seven species (T. asymmetrica, T. andina, T. eichleriana ECU6, T. eichleriana ECU4 T. pinicola, T. violea), supported by bootstrap values from 72% to 100%. From the 2D secondary structure alignment, three types of consensus models with helices and loops were obtained. Thus, T. albida belongs to type I; T. eichleriana, T. tomaculum, and T. violea belong to type II; and T. asymmetrica, T. andina, T. pinicola, and T. spp. (GER) belong to type III; each type contains four to six domains, with nine CBCs among these that corroborate different species.
Keywords: ITS2; alignment-based; alignment-free; compensatory base changes; secondary structure; species delimitation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chethana K.W., Manawasinghe I.S., Hurdeal V.G., Bhunjun C.S., Appadoo M.A., Gentekaki E., Raspé O., Promputtha I., Hyde K.D. What are fungal species and how to delineate them? Fungal Divers. 2021;109:1–25. doi: 10.1007/s13225-021-00483-9. - DOI
-
- Ding X., Xiao J.H., Li L., Conran J.G., Li J. Congruent species delimitation of two controversial gold-thread nanmu tree species based on morphological and restriction site-associated DNA sequencing data. J. Syst. Evol. 2019;57:234–246. doi: 10.1111/jse.12433. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

