Calling Cards: A Customizable Platform to Longitudinally Record Protein-DNA Interactions Over Time in Cells and Tissues
- PMID: 37755132
- PMCID: PMC10627244
- DOI: 10.1002/cpz1.883
Calling Cards: A Customizable Platform to Longitudinally Record Protein-DNA Interactions Over Time in Cells and Tissues
Erratum in
-
Correction: Calling Cards: A Customizable Platform to Longitudinally Record Protein-DNA Interactions Over Time in Cells and Tissues.Curr Protoc. 2023 Oct;3(10):e928. doi: 10.1002/cpz1.928. Curr Protoc. 2023. PMID: 37855731 No abstract available.
Abstract
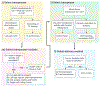
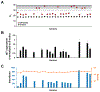
Calling Cards is a platform technology to record a cumulative history of transient protein-DNA interactions in the genome of genetically targeted cell types. The record of these interactions is recovered by next-generation sequencing. Compared with other genomic assays, readouts of which provide a snapshot at the time of harvest, Calling Cards enables correlation of historical molecular states to eventual outcomes or phenotypes. To achieve this, Calling Cards uses the piggyBac transposase to insert self-reporting transposon "Calling Cards" into the genome, leaving permanent marks at interaction sites. Calling Cards can be deployed in a variety of in vitro and in vivo biological systems to study gene regulatory networks involved in development, aging, and disease. Out of the box, it assesses enhancer usage but can be adapted to profile-specific transcription factor (TF) binding with custom TF-piggyBac fusion proteins. The Calling Cards workflow has five main stages: delivery of Calling Cards reagents, sample preparation, library preparation, sequencing, and data analysis. Here, we first present a comprehensive guide for experimental design, reagent selection, and optional customization of the platform to study additional TFs. Then, we provide an updated protocol for the five steps, using reagents that improve throughput and decrease costs, including an overview of a newly deployed computational pipeline. This protocol is designed for users with basic molecular biology experience to process samples into sequencing libraries in 2 days. Familiarity with bioinformatic analysis and command line tools is required to set up the pipeline in a high-performance computing environment and to conduct downstream analyses. © 2023 Wiley Periodicals LLC. Basic Protocol 1: Preparation and delivery of Calling Cards reagents Support Protocol 1: Next-generation sequencing quantification of barcode distribution within self-reporting transposon plasmid pool and adeno-associated virus genome Basic Protocol 2: Sample collection and RNA purification Support Protocol 2: Library density quantitative PCR Basic Protocol 3: Sequencing library preparation Basic Protocol 4: Library pooling and sequencing Basic Protocol 5: Data analysis.
Keywords: calling cards; enhancer usage; epigenetics; gene regulation; transcription factor binding.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
RDM and AM are listed as inventors on US patent US20200181626A1.
Figures









Update of
-
Calling Cards: a customizable platform to longitudinally record protein-DNA interactions over time in cells and tissues.bioRxiv [Preprint]. 2023 Jun 9:2023.06.07.544098. doi: 10.1101/2023.06.07.544098. bioRxiv. 2023. Update in: Curr Protoc. 2023 Sep;3(9):e883. doi: 10.1002/cpz1.883. PMID: 37333130 Free PMC article. Updated. Preprint.
References
-
- Challis RC, Ravindra Kumar S, Chan KY, Challis C, Beadle K, Jang MJ, Kim HM, Rajendran PS, Tompkins JD, Shivkumar K, et al. 2019. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nature Protocols 14:379–414. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

