Novel marine metalloprotease-new approaches for inhibition of biofilm formation of Stenotrophomonas maltophilia
- PMID: 37755512
- PMCID: PMC10638167
- DOI: 10.1007/s00253-023-12781-0
Novel marine metalloprotease-new approaches for inhibition of biofilm formation of Stenotrophomonas maltophilia
Abstract
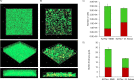
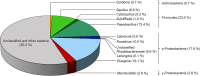
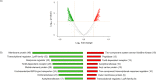
Many marine organisms produce bioactive molecules with unique characteristics to survive in their ecological niches. These enzymes can be applied in biotechnological processes and in the medical sector to replace aggressive chemicals that are harmful to the environment. Especially in the human health sector, there is a need for new approaches to fight against pathogens like Stenotrophomonas maltophilia which forms thick biofilms on artificial joints or catheters and causes serious diseases. Our approach was to use enrichment cultures of five marine resources that underwent sequence-based screenings in combination with deep omics analyses in order to identify enzymes with antibiofilm characteristics. Especially the supernatant of the enrichment culture of a stony coral caused a 40% reduction of S. maltophilia biofilm formation. In the presence of the supernatant, our transcriptome dataset showed a clear stress response (upregulation of transcripts for metal resistance, antitoxins, transporter, and iron acquisition) to the treatment. Further investigation of the enrichment culture metagenome and proteome indicated a series of potential antimicrobial enzymes. We found an impressive group of metalloproteases in the proteome of the supernatant that is responsible for the detected anti-biofilm effect against S. maltophilia. KEY POINTS: • Omics-based discovery of novel marine-derived antimicrobials for human health management by inhibition of S. maltophilia • Up to 40% reduction of S. maltophilia biofilm formation by the use of marine-derived samples • Metalloprotease candidates prevent biofilm formation of S. maltophilia K279a by up to 20.
Keywords: Antimicrobials; Human health management; Marine habitats; Proteases; Stenotrophomonas maltophilia.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Abda EM, Krysciak D, Krohn-Molt I, Mamat U, Schmeisser C, Forstner KU, Schaible UE, Kohl TA, Nieman S, Streit WR. Phenotypic heterogeneity affects Stenotrophomonas maltophilia K279a colony morphotypes and beta-lactamase expression. Front Microbiol. 2015;6:1373. doi: 10.3389/fmicb.2015.01373. - DOI - PMC - PubMed
-
- Abfalter CM, Schönauer E, Ponnuraj K, Huemer M, Gadermaier G, Regl C, Briza P, Ferreira F, Huber CG, Brandstetter H, Posselt G, Wessler S. Cloning, purification and characterization of the collagenase ColA expressed by Bacillus cereus ATCC 14579. PLoS ONE. 2016;11:e0162433. doi: 10.1371/journal.pone.0162433. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

