Lymphoma in Border Collies: Genome-Wide Association and Pedigree Analysis
- PMID: 37756103
- PMCID: PMC10536503
- DOI: 10.3390/vetsci10090581
Lymphoma in Border Collies: Genome-Wide Association and Pedigree Analysis
Abstract
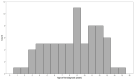
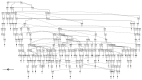
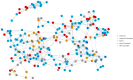
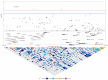
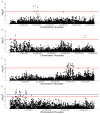
There has been considerable interest in studying cancer in dogs and its potential as a model system for humans. One area of research has been the search for genetic risk variants in canine lymphoma, which is amongst the most common canine cancers. Previous studies have focused on a limited number of breeds, but none have included Border Collies. The aims of this study were to identify relationships between Border Collie lymphoma cases through an extensive pedigree investigation and to utilise relationship information to conduct genome-wide association study (GWAS) analyses to identify risk regions associated with lymphoma. The expanded pedigree analysis included 83,000 Border Collies, with 71 identified lymphoma cases. The analysis identified affected close relatives, and a common ancestor was identified for 54 cases. For the genomic study, a GWAS was designed to incorporate lymphoma cases, putative "carriers", and controls. A case-control GWAS was also conducted as a comparison. Both analyses showed significant SNPs in regions on chromosomes 18 and 27. Putative top candidate genes from these regions included DLA-79, WNT10B, LMBR1L, KMT2D, and CCNT1.
Keywords: Border Collie; GWAS; breed; cancer; canine; dogs; lymphoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Comazzi S., Marelli S., Cozzi M., Rizzi R., Finotello R., Henriques J., Pastor J., Ponce F., Rohrer-Bley C., Rutgen B.C., et al. Breed-associated risks for developing canine lymphoma differ among countries: An European canine lymphoma network study. BMC Vet. Res. 2018;14:232. doi: 10.1186/s12917-018-1557-2. - DOI - PMC - PubMed
-
- Modiano J.F., Breen M., Burnett R.C., Parker H.G., Inusah S., Thomas R., Avery P.R., Lindblad-Toh K., Ostrander E.A., Cutter G.C., et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

