Cell state transition analysis identifies interventions that improve control of Mycobacterium tuberculosis infection by susceptible macrophages
- PMID: 37756395
- PMCID: PMC10530096
- DOI: 10.1126/sciadv.adh4119
Cell state transition analysis identifies interventions that improve control of Mycobacterium tuberculosis infection by susceptible macrophages
Abstract
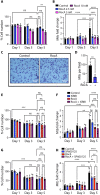
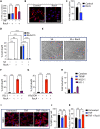
Understanding cell state transitions and purposefully controlling them to improve therapies is a longstanding challenge in biological research and medicine. Here, we identify a transcriptional signature that distinguishes activated macrophages from the tuberculosis (TB) susceptible and resistant mice. We then apply the cSTAR (cell state transition assessment and regulation) approach to data from screening-by-RNA sequencing to identify chemical perturbations that shift the transcriptional state of tumor necrosis factor (TNF)-activated TB-susceptible macrophages toward that of TB-resistant cells, i.e., prevents their aberrant activation without suppressing beneficial TNF responses. Last, we demonstrate that the compounds identified with this approach enhance the resistance of the TB-susceptible mouse macrophages to virulent Mycobacterium tuberculosis.
Figures



Update of
-
Cell state transition analysis identifies interventions that improve control of M. tuberculosis infection by susceptible macrophages.bioRxiv [Preprint]. 2023 Feb 10:2023.02.09.527908. doi: 10.1101/2023.02.09.527908. bioRxiv. 2023. Update in: Sci Adv. 2023 Sep 29;9(39):eadh4119. doi: 10.1126/sciadv.adh4119. PMID: 36798271 Free PMC article. Updated. Preprint.
Similar articles
-
Cell state transition analysis identifies interventions that improve control of M. tuberculosis infection by susceptible macrophages.bioRxiv [Preprint]. 2023 Feb 10:2023.02.09.527908. doi: 10.1101/2023.02.09.527908. bioRxiv. 2023. Update in: Sci Adv. 2023 Sep 29;9(39):eadh4119. doi: 10.1126/sciadv.adh4119. PMID: 36798271 Free PMC article. Updated. Preprint.
-
Targeting Mycobacterium tuberculosis Tumor Necrosis Factor Alpha-Downregulating Genes for the Development of Antituberculous Vaccines.mBio. 2016 May 31;7(3):e01023-15. doi: 10.1128/mBio.01023-15. mBio. 2016. PMID: 27247233 Free PMC article.
-
Small Molecule Mediated Restoration of Mitochondrial Function Augments Anti-Mycobacterial Activity of Human Macrophages Subjected to Cholesterol Induced Asymptomatic Dyslipidemia.Front Cell Infect Microbiol. 2017 Oct 10;7:439. doi: 10.3389/fcimb.2017.00439. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29067283 Free PMC article.
-
Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies.Semin Immunopathol. 2016 Mar;38(2):221-37. doi: 10.1007/s00281-015-0538-9. Epub 2015 Nov 5. Semin Immunopathol. 2016. PMID: 26542392 Free PMC article. Review.
-
Cell death at the cross roads of host-pathogen interaction in Mycobacterium tuberculosis infection.Tuberculosis (Edinb). 2018 Dec;113:99-121. doi: 10.1016/j.tube.2018.09.007. Epub 2018 Sep 29. Tuberculosis (Edinb). 2018. PMID: 30514519 Review.
Cited by
-
cSTAR analysis identifies endothelial cell cycle as a key regulator of flow-dependent artery remodeling.Sci Adv. 2025 Jan 3;11(1):eado9970. doi: 10.1126/sciadv.ado9970. Epub 2025 Jan 3. Sci Adv. 2025. PMID: 39752487 Free PMC article.
-
Cell State Transition Models Stratify Breast Cancer Cell Phenotypes and Reveal New Therapeutic Targets.Cancers (Basel). 2024 Jun 27;16(13):2354. doi: 10.3390/cancers16132354. Cancers (Basel). 2024. PMID: 39001416 Free PMC article.
-
Lipid Peroxidation and Type I Interferon Coupling Fuels Pathogenic Macrophage Activation Causing Tuberculosis Susceptibility.bioRxiv [Preprint]. 2025 May 1:2024.03.05.583602. doi: 10.1101/2024.03.05.583602. bioRxiv. 2025. PMID: 38496444 Free PMC article. Preprint.
-
Comparative transcriptomic analysis of mouse macrophages infected with live attenuated vaccine strains of Mycobacterium tuberculosis.Front Immunol. 2025 Jul 11;16:1583439. doi: 10.3389/fimmu.2025.1583439. eCollection 2025. Front Immunol. 2025. PMID: 40718493 Free PMC article.
References
-
- D. Fey, M. Halasz, D. Dreidax, S. P. Kennedy, J. F. Hastings, N. Rauch, A. G. Munoz, R. Pilkington, M. Fischer, F. Westermann, W. Kolch, B. N. Kholodenko, D. R. Croucher, Signaling pathway models as biomarkers: Patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci. Signal. 8, ra130 (2015). - PubMed
-
- Y. Su, M. E. Ko, H. Cheng, R. Zhu, M. Xue, J. Wang, J. W. Lee, L. Frankiw, A. Xu, S. Wong, L. Robert, K. Takata, D. Yuan, Y. Lu, S. Huang, A. Ribas, R. Levine, G. P. Nolan, W. Wei, S. K. Plevritis, G. Li, D. Baltimore, J. R. Heath, Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat. Commun. 11, 2345 (2020). - PMC - PubMed
-
- X. Qiu, Y. Zhang, J. D. Martin-Rufino, C. Weng, S. Hosseinzadeh, D. Yang, A. N. Pogson, M. Y. Hein, K. Hoi Min, L. Wang, E. I. Grody, M. J. Shurtleff, R. Yuan, S. Xu, Y. Ma, J. M. Replogle, E. S. Lander, S. Darmanis, I. Bahar, V. G. Sankaran, J. Xing, J. S. Weissman, Mapping transcriptomic vector fields of single cells. Cell 185, 690–711.e45 (2022). - PMC - PubMed
-
- A. R. DiNardo, T. Gandhi, J. Heyckendorf, S. L. Grimm, K. Rajapakshe, T. Nishiguchi, M. Reimann, H. L. Kirchner, J. Kahari, Q. Dlamini, C. Lange, T. Goldmann, S. Marwitz; DZIF-TB cohort study group, A. Abhimanyu, J. D. Cirillo, S. H. E. Kaufmann, M. G. Netea, R. V. Crevel, A. M. Mandalakas, C. Coarfa, Gene expression signatures identify biologically and clinically distinct tuberculosis endotypes. European Respir. J. 60, 2102263 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

