Activation of CD8+ T Cells in Chronic Obstructive Pulmonary Disease Lung
- PMID: 37756440
- PMCID: PMC10868372
- DOI: 10.1164/rccm.202305-0924OC
Activation of CD8+ T Cells in Chronic Obstructive Pulmonary Disease Lung
Abstract
Rationale: Despite the importance of inflammation in chronic obstructive pulmonary disease (COPD), the immune cell landscape in the lung tissue of patients with mild-moderate disease has not been well characterized at the single-cell and molecular level. Objectives: To define the immune cell landscape in lung tissue from patients with mild-moderate COPD at single-cell resolution. Methods: We performed single-cell transcriptomic, proteomic, and T-cell receptor repertoire analyses on lung tissue from patients with mild-moderate COPD (n = 5, Global Initiative for Chronic Obstructive Lung Disease I or II), emphysema without airflow obstruction (n = 5), end-stage COPD (n = 2), control (n = 6), or donors (n = 4). We validated in an independent patient cohort (N = 929) and integrated with the Hhip+/- murine model of COPD. Measurements and Main Results: Mild-moderate COPD lungs have increased abundance of two CD8+ T cell subpopulations: cytotoxic KLRG1+TIGIT+CX3CR1+ TEMRA (T effector memory CD45RA+) cells, and DNAM-1+CCR5+ T resident memory (TRM) cells. These CD8+ T cells interact with myeloid and alveolar type II cells via IFNG and have hyperexpanded T-cell receptor clonotypes. In an independent cohort, the CD8+KLRG1+ TEMRA cells are increased in mild-moderate COPD lung compared with control or end-stage COPD lung. Human CD8+KLRG1+ TEMRA cells are similar to CD8+ T cells driving inflammation in an aging-related murine model of COPD. Conclusions: CD8+ TEMRA cells are increased in mild-moderate COPD lung and may contribute to inflammation that precedes severe disease. Further study of these CD8+ T cells may have therapeutic implications for preventing severe COPD.
Keywords: RNA sequence analysis; chronic obstructive pulmonary disease; memory T cells; multiomics; proteomics.
Figures


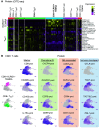





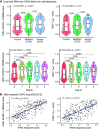

Comment in
-
Bad Neighbors or Bad Neighborhoods: Pathogenic Residency of T Cells in Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2023 Dec 1;208(11):1148-1150. doi: 10.1164/rccm.202310-1760ED. Am J Respir Crit Care Med. 2023. PMID: 37855741 Free PMC article. No abstract available.
References
-
- Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. - PubMed
-
- Barnes PJ. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2016;138:16–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

