Germ line variant GFI1-36N affects DNA repair and sensitizes AML cells to DNA damage and repair therapy
- PMID: 37756525
- PMCID: PMC10733838
- DOI: 10.1182/blood.2022015752
Germ line variant GFI1-36N affects DNA repair and sensitizes AML cells to DNA damage and repair therapy
Abstract
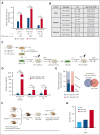
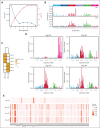
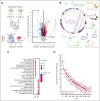
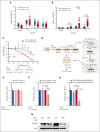
Growth factor independence 1 (GFI1) is a DNA-binding transcription factor and a key regulator of hematopoiesis. GFI1-36N is a germ line variant, causing a change of serine (S) to asparagine (N) at position 36. We previously reported that the GFI1-36N allele has a prevalence of 10% to 15% among patients with acute myeloid leukemia (AML) and 5% to 7% among healthy Caucasians and promotes the development of this disease. Using a multiomics approach, we show here that GFI1-36N expression is associated with increased frequencies of chromosomal aberrations, mutational burden, and mutational signatures in both murine and human AML and impedes homologous recombination (HR)-directed DNA repair in leukemic cells. GFI1-36N exhibits impaired binding to N-Myc downstream-regulated gene 1 (Ndrg1) regulatory elements, causing decreased NDRG1 levels, which leads to a reduction of O6-methylguanine-DNA-methyltransferase (MGMT) expression levels, as illustrated by both transcriptome and proteome analyses. Targeting MGMT via temozolomide, a DNA alkylating drug, and HR via olaparib, a poly-ADP ribose polymerase 1 inhibitor, caused synthetic lethality in human and murine AML samples expressing GFI1-36N, whereas the effects were insignificant in nonmalignant GFI1-36S or GFI1-36N cells. In addition, mice that received transplantation with GFI1-36N leukemic cells treated with a combination of temozolomide and olaparib had significantly longer AML-free survival than mice that received transplantation with GFI1-36S leukemic cells. This suggests that reduced MGMT expression leaves GFI1-36N leukemic cells particularly vulnerable to DNA damage initiating chemotherapeutics. Our data provide critical insights into novel options to treat patients with AML carrying the GFI1-36N variant.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: H.C.R. received consulting and lecture fees from AbbVie, AstraZeneca, Vertex, and Merck; received research funding from Gilead Pharmaceuticals; and is a cofounder of CDL Therapeutics GmbH. G.H. received consulting and lecture fees from Novartis, Incyte, and Jazz Pharmaceuticals. C.K. received funding from AstraZeneca for this project. E.W. received funding from Pfizer for conducting the clinical trial. The remaining authors declare no competing financial interests.
B.O. and U.D. are retired from University Hospital Essen, Essen, Germany.
Figures





References
-
- Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–1007. - PubMed
-
- Karsunky H, Zeng H, Schmidt T, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30(3):295–300. - PubMed
-
- Thambyrajah R, Mazan M, Patel R, et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. 2016;18(1):21–32. - PubMed
-
- Yucel R, Kosan C, Heyd F, Moroy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem. 2004;279(39):40906–40917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

