A normative microbiome is not restored following kidney transplantation
- PMID: 37756543
- PMCID: PMC10582644
- DOI: 10.1042/CS20230779
A normative microbiome is not restored following kidney transplantation
Abstract
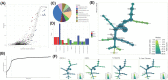
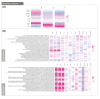
Dialysis and kidney transplantation (Ktx) mitigate some of the physiological deficits in chronic kidney disease (CKD), but it remains to be determined if these mitigate microbial dysbiosis and the production of inflammatory microbial metabolites, which contribute significantly to the uraemic phenotype. We have investigated bacterial DNA signatures present in the circulation of CKD patients and those receiving a KTx. Our data are consistent with increasing dysbiosis as CKD progresses, with an accompanying increase in trimethylamine (TMA) producing pathobionts Pseudomonas and Bacillus. Notably, KTx patients displayed a significantly different microbiota compared with CKD5 patients, which surprisingly included further increase in TMA producing Bacillus and loss of salutogenic Lactobacilli. Only two genera (Viellonella and Saccharimonidales) showed significant differences in abundance following KTx that may reflect a reciprocal relationship between TMA producers and utilisers, which supersedes restoration of a normative microbiome. Our metadata analysis confirmed that TMA N-oxide (TMAO) along with one carbon metabolism had significant impact upon both inflammatory burden and the composition of the microbiome. This indicates that these metabolites are key to shaping the uraemic microbiome and might be exploited in the development of dietary intervention strategies to both mitigate the physiological deficits in CKD and enable the restoration of a more salutogenic microbiome.
Keywords: chronic kidney disease; kidney transplantation; microbiome.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



Similar articles
-
Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients.Sci Rep. 2017 May 3;7(1):1445. doi: 10.1038/s41598-017-01387-y. Sci Rep. 2017. PMID: 28469156 Free PMC article.
-
Characterizing the gut microbiota in patients with chronic kidney disease.Postgrad Med. 2020 Aug;132(6):495-505. doi: 10.1080/00325481.2020.1744335. Epub 2020 Apr 2. Postgrad Med. 2020. PMID: 32241215
-
Gut Microbiota in Patients Receiving Dialysis: A Review.Pathogens. 2024 Sep 15;13(9):801. doi: 10.3390/pathogens13090801. Pathogens. 2024. PMID: 39338992 Free PMC article. Review.
-
The Relationship between Choline Bioavailability from Diet, Intestinal Microbiota Composition, and Its Modulation of Human Diseases.Nutrients. 2020 Aug 5;12(8):2340. doi: 10.3390/nu12082340. Nutrients. 2020. PMID: 32764281 Free PMC article. Review.
-
Gut microbiome in chronic kidney disease.Exp Physiol. 2016 Apr;101(4):471-7. doi: 10.1113/EP085283. Epub 2015 Oct 2. Exp Physiol. 2016. PMID: 26337794 Review.
Cited by
-
Sweet, bloody consumption - what we eat and how it affects vascular ageing, the BBB and kidney health in CKD.Gut Microbes. 2024 Jan-Dec;16(1):2341449. doi: 10.1080/19490976.2024.2341449. Epub 2024 Apr 30. Gut Microbes. 2024. PMID: 38686499 Free PMC article. Review.

