Transcription inhibition suppresses nuclear blebbing and rupture independently of nuclear rigidity
- PMID: 37756607
- PMCID: PMC10660790
- DOI: 10.1242/jcs.261547
Transcription inhibition suppresses nuclear blebbing and rupture independently of nuclear rigidity
Abstract
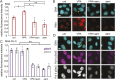
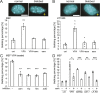
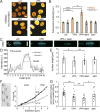
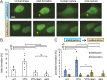
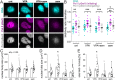
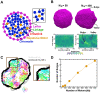
Chromatin plays an essential role in the nuclear mechanical response and determining nuclear shape, which maintain nuclear compartmentalization and function. However, major genomic functions, such as transcription activity, might also impact cell nuclear shape via blebbing and rupture through their effects on chromatin structure and dynamics. To test this idea, we inhibited transcription with several RNA polymerase II inhibitors in wild-type cells and perturbed cells that presented increased nuclear blebbing. Transcription inhibition suppressed nuclear blebbing for several cell types, nuclear perturbations and transcription inhibitors. Furthermore, transcription inhibition suppressed nuclear bleb formation, bleb stabilization and bleb-based nuclear ruptures. Interestingly, transcription inhibition did not alter the histone H3 lysine 9 (H3K9) modification state, nuclear rigidity, and actin compression and contraction, which typically control nuclear blebbing. Polymer simulations suggested that RNA polymerase II motor activity within chromatin could drive chromatin motions that deform the nuclear periphery. Our data provide evidence that transcription inhibition suppresses nuclear blebbing and rupture, in a manner separate and distinct from chromatin rigidity.
Keywords: Chromatin; Mechanobiology; Nuclear blebbing; Nucleus.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
-
- Banigan, E. J., Tang, W., van den Berg, A. A., Stocsits, R. R., Wutz, G., Brandão, H. B., Busslinger, G. A., Peters, J.-M. and Mirny, L. A. (2023). Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proc. Natl. Acad. Sci. USA 120, e2210480120. 10.1073/pnas.2210480120 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

