Viscosity Prediction of High-Concentration Antibody Solutions with Atomistic Simulations
- PMID: 37757589
- PMCID: PMC10565822
- DOI: 10.1021/acs.jcim.3c00947
Viscosity Prediction of High-Concentration Antibody Solutions with Atomistic Simulations
Abstract
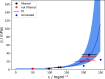
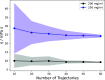
The computational prediction of the viscosity of dense protein solutions is highly desirable, for example, in the early development phase of high-concentration biopharmaceutical formulations where the material needed for experimental determination is typically limited. Here, we use large-scale atomistic molecular dynamics (MD) simulations with explicit solvation to de novo predict the dynamic viscosities of solutions of a monoclonal IgG1 antibody (mAb) from the pressure fluctuations using a Green-Kubo approach. The viscosities at simulated mAb concentrations of 200 and 250 mg/mL are compared to the experimental values, which we measured with rotational rheometry. The computational viscosity of 24 mPa·s at the mAb concentration of 250 mg/mL matches the experimental value of 23 mPa·s obtained at a concentration of 213 mg/mL, indicating slightly different effective concentrations (or activities) in the MD simulations and in the experiments. This difference is assigned to a slight underestimation of the effective mAb-mAb interactions in the simulations, leading to a too loose dynamic mAb network that governs the viscosity. Taken together, this study demonstrates the feasibility of all-atom MD simulations for predicting the properties of dense mAb solutions and provides detailed microscopic insights into the underlying molecular interactions. At the same time, it also shows that there is room for further improvements and highlights challenges, such as the massive sampling required for computing collective properties of dense biomolecular solutions in the high-viscosity regime with reasonable statistical precision.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
LinkOut - more resources
Full Text Sources

