Unmodificated stepless regulation of CRISPR/Cas12a multi-performance
- PMID: 37757856
- PMCID: PMC10602922
- DOI: 10.1093/nar/gkad748
Unmodificated stepless regulation of CRISPR/Cas12a multi-performance
Abstract
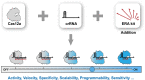
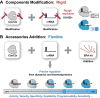
As CRISPR technology is promoted to more fine-divided molecular biology applications, its inherent performance finds it increasingly difficult to cope with diverse needs in these different fields, and how to more accurately control the performance has become a key issue to develop CRISPR technology to a new stage. Herein, we propose a CRISPR/Cas12a regulation strategy based on the powerful programmability of nucleic acid nanotechnology. Unlike previous difficult and rigid regulation of core components Cas nuclease and crRNA, only a simple switch of different external RNA accessories is required to change the reaction kinetics or thermodynamics, thereby finely and almost steplessly regulating multi-performance of CRISPR/Cas12a including activity, speed, specificity, compatibility, programmability and sensitivity. In particular, the significantly improved specificity is expected to mark advance the accuracy of molecular detection and the safety of gene editing. In addition, this strategy was applied to regulate the delayed activation of Cas12a, overcoming the compatibility problem of the one-pot assay without any physical separation or external stimulation, and demonstrating great potential for fine-grained control of CRISPR. This simple but powerful CRISPR regulation strategy without any component modification has pioneering flexibility and versatility, and will unlock the potential for deeper applications of CRISPR technology in many finely divided fields.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Harnessing noncanonical crRNAs to improve functionality of Cas12a orthologs.Cell Rep. 2024 Feb 27;43(2):113777. doi: 10.1016/j.celrep.2024.113777. Epub 2024 Feb 14. Cell Rep. 2024. PMID: 38358883 Free PMC article.
-
Universal crRNA Acylation Strategy for Robust Photo-Initiated One-Pot CRISPR-Cas12a Nucleic Acid Diagnostics.Angew Chem Int Ed Engl. 2024 Jun 3;63(23):e202401486. doi: 10.1002/anie.202401486. Epub 2024 Apr 30. Angew Chem Int Ed Engl. 2024. PMID: 38563640
-
Light-Start CRISPR-Cas12a Reaction with Caged crRNA Enables Rapid and Sensitive Nucleic Acid Detection.Angew Chem Int Ed Engl. 2023 Jun 5;62(23):e202300663. doi: 10.1002/anie.202300663. Epub 2023 Apr 27. Angew Chem Int Ed Engl. 2023. PMID: 37016515
-
CRISPR-Cas12a System for Biosensing and Gene Regulation.Chem Asian J. 2021 Apr 19;16(8):857-867. doi: 10.1002/asia.202100043. Epub 2021 Mar 18. Chem Asian J. 2021. PMID: 33638271 Review.
-
Application of CRISPR/Cas12a in the rapid detection of pathogens.Clin Chim Acta. 2023 Aug 1;548:117520. doi: 10.1016/j.cca.2023.117520. Epub 2023 Aug 16. Clin Chim Acta. 2023. PMID: 37595863 Review.
Cited by
-
Nonequilibrium hybridization-driven CRISPR/Cas adapter with extended energetic penalty for discrimination of single-nucleotide variants.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf287. doi: 10.1093/nar/gkaf287. Nucleic Acids Res. 2025. PMID: 40243059 Free PMC article.
-
Modular CRISPR/Cas12a synergistic activation platform for detection and logic operations.Nucleic Acids Res. 2024 Jul 8;52(12):7384-7396. doi: 10.1093/nar/gkae470. Nucleic Acids Res. 2024. PMID: 38828769 Free PMC article.
-
Synthetic CRISPR Networks Driven by Transcription Factors via Structure-Switching DNA Translators.J Am Chem Soc. 2025 Jun 18;147(24):21184-21193. doi: 10.1021/jacs.5c06913. Epub 2025 Jun 9. J Am Chem Soc. 2025. PMID: 40491004 Free PMC article.
-
Regulation of CRISPR trans-cleavage activity by an overhanging activator.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf117. doi: 10.1093/nar/gkaf117. Nucleic Acids Res. 2025. PMID: 39995038 Free PMC article.
-
THRUST: translesion synthesis-driven hierarchical regulation using a template-activator construct for Cas12a activity.Chem Sci. 2025 Jun 16;16(28):12917-12926. doi: 10.1039/d5sc02575c. eCollection 2025 Jul 16. Chem Sci. 2025. PMID: 40529559 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

