Proteome census upon nutrient stress reveals Golgiphagy membrane receptors
- PMID: 37757899
- PMCID: PMC10620096
- DOI: 10.1038/s41586-023-06657-6
Proteome census upon nutrient stress reveals Golgiphagy membrane receptors
Abstract
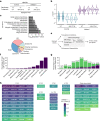
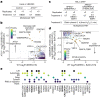
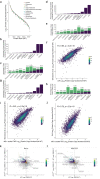
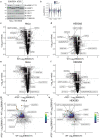
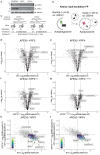
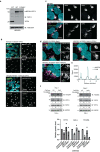
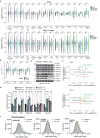
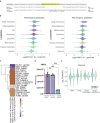
During nutrient stress, macroautophagy degrades cellular macromolecules, thereby providing biosynthetic building blocks while simultaneously remodelling the proteome1,2. Although the machinery responsible for initiation of macroautophagy has been well characterized3,4, our understanding of the extent to which individual proteins, protein complexes and organelles are selected for autophagic degradation, and the underlying targeting mechanisms, is limited. Here we use orthogonal proteomic strategies to provide a spatial proteome census of autophagic cargo during nutrient stress in mammalian cells. We find that macroautophagy has selectivity for recycling membrane-bound organelles (principally Golgi and endoplasmic reticulum). Through autophagic cargo prioritization, we identify a complex of membrane-embedded proteins, YIPF3 and YIPF4, as receptors for Golgiphagy. During nutrient stress, YIPF3 and YIPF4 interact with ATG8 proteins through LIR motifs and are mobilized into autophagosomes that traffic to lysosomes in a process that requires the canonical autophagic machinery. Cells lacking YIPF3 or YIPF4 are selectively defective in elimination of a specific cohort of Golgi membrane proteins during nutrient stress. Moreover, YIPF3 and YIPF4 play an analogous role in Golgi remodelling during programmed conversion of stem cells to the neuronal lineage in vitro. Collectively, the findings of this study reveal prioritization of membrane protein cargo during nutrient-stress-dependent proteome remodelling and identify a Golgi remodelling pathway that requires membrane-embedded receptors.
© 2023. The Author(s).
Conflict of interest statement
J.W.H. is a founder and consultant for Caraway Therapeutics and a co-founding board member of Interline Therapeutics. All other authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

