Nutrient-dependent signaling pathways that control autophagy in yeast
- PMID: 37758520
- PMCID: PMC10841420
- DOI: 10.1002/1873-3468.14741
Nutrient-dependent signaling pathways that control autophagy in yeast
Abstract
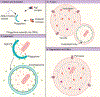
Macroautophagy/autophagy is a highly conserved catabolic process vital for cellular stress responses and maintaining equilibrium within the cell. Malfunctioning autophagy has been implicated in the pathogenesis of various diseases, including certain neurodegenerative disorders, diabetes, metabolic diseases, and cancer. Cells face diverse metabolic challenges, such as limitations in nitrogen, carbon, and minerals such as phosphate and iron, necessitating the integration of complex metabolic information. Cells utilize a signal transduction network of sensors, transducers, and effectors to coordinate the execution of the autophagic response, concomitant with the severity of the nutrient-starvation condition. This review presents the current mechanistic understanding of how cells regulate the initiation of autophagy through various nutrient-dependent signaling pathways. Emphasizing findings from studies in yeast, we explore the emerging principles that underlie the nutrient-dependent regulation of autophagy, significantly shaping stress-induced autophagy responses under various metabolic stress conditions.
Keywords: AMPK; PKA; TOR; autophagy regulation; nutrient homeostasis; signaling.
© 2023 Federation of European Biochemical Societies.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

