piRNA processing by a trimeric Schlafen-domain nuclease
- PMID: 37758951
- PMCID: PMC10567574
- DOI: 10.1038/s41586-023-06588-2
piRNA processing by a trimeric Schlafen-domain nuclease
Erratum in
-
Author Correction: piRNA processing by a trimeric Schlafen-domain nuclease.Nature. 2024 Aug;632(8027):E31. doi: 10.1038/s41586-024-07938-4. Nature. 2024. PMID: 39143225 Free PMC article. No abstract available.
Abstract
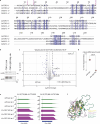
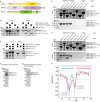
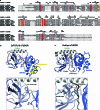
Transposable elements are genomic parasites that expand within and spread between genomes1. PIWI proteins control transposon activity, notably in the germline2,3. These proteins recognize their targets through small RNA co-factors named PIWI-interacting RNAs (piRNAs), making piRNA biogenesis a key specificity-determining step in this crucial genome immunity system. Although the processing of piRNA precursors is an essential step in this process, many of the molecular details remain unclear. Here, we identify an endoribonuclease, precursor of 21U RNA 5'-end cleavage holoenzyme (PUCH), that initiates piRNA processing in the nematode Caenorhabditis elegans. Genetic and biochemical studies show that PUCH, a trimer of Schlafen-like-domain proteins (SLFL proteins), executes 5'-end piRNA precursor cleavage. PUCH-mediated processing strictly requires a 7-methyl-G cap (m7G-cap) and a uracil at position three. We also demonstrate how PUCH interacts with PETISCO, a complex that binds to piRNA precursors4, and that this interaction enhances piRNA production in vivo. The identification of PUCH concludes the search for the 5'-end piRNA biogenesis factor in C. elegans and uncovers a type of RNA endonuclease formed by three SLFL proteins. Mammalian Schlafen (SLFN) genes have been associated with immunity5, exposing a molecular link between immune responses in mammals and deeply conserved RNA-based mechanisms that control transposable elements.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

