Defining super-enhancers by highly ranked histone H4 multi-acetylation levels identifies transcription factors associated with glioblastoma stem-like properties
- PMID: 37759202
- PMCID: PMC10523799
- DOI: 10.1186/s12864-023-09659-w
Defining super-enhancers by highly ranked histone H4 multi-acetylation levels identifies transcription factors associated with glioblastoma stem-like properties
Abstract
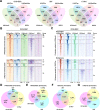
Background: Super-enhancers (SEs), which activate genes involved in cell-type specificity, have mainly been defined as genomic regions with top-ranked enrichment(s) of histone H3 with acetylated K27 (H3K27ac) and/or transcription coactivator(s) including a bromodomain and extra-terminal domain (BET) family protein, BRD4. However, BRD4 preferentially binds to multi-acetylated histone H4, typically with acetylated K5 and K8 (H4K5acK8ac), leading us to hypothesize that SEs should be defined by high H4K5acK8ac enrichment at least as well as by that of H3K27ac.
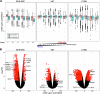
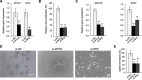
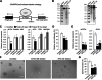
Results: Here, we conducted genome-wide profiling of H4K5acK8ac and H3K27ac, BRD4 binding, and the transcriptome by using a BET inhibitor, JQ1, in three human glial cell lines. When SEs were defined as having the top ranks for H4K5acK8ac or H3K27ac signal, 43% of H4K5acK8ac-ranked SEs were distinct from H3K27ac-ranked SEs in a glioblastoma stem-like cell (GSC) line. CRISPR-Cas9-mediated deletion of the H4K5acK8ac-preferred SEs associated with MYCN and NFIC decreased the stem-like properties in GSCs.
Conclusions: Collectively, our data highlights H4K5acK8ac's utility for identifying genes regulating cell-type specificity.
Keywords: Epigenetics; Histone acetylation; Inhibitor; Nucleosome; Tumorigenesis.
© 2023. The Author(s).
Conflict of interest statement
BK took a position with AstraZeneca R&D during the submission of this manuscript. AstraZeneca R&D was not involved at any stage of the work presented here and there is no conflict of interest related to AstraZeneca R&D for this work. BK may own stock options of AstraZeneca. The authors declare no competing interests.
Figures



References
-
- Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol. 2014;15:723–734. - PubMed
-
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. - PubMed
-
- Kim S, Shendure J. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell. 2019;76:306–319. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

