Inhibition of extracellular vesicle-derived miR-146a-5p decreases progression of melanoma brain metastasis via Notch pathway dysregulation in astrocytes
- PMID: 37759347
- PMCID: PMC10533779
- DOI: 10.1002/jev2.12363
Inhibition of extracellular vesicle-derived miR-146a-5p decreases progression of melanoma brain metastasis via Notch pathway dysregulation in astrocytes
Abstract
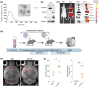
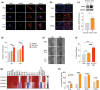
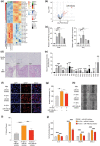
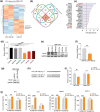
Melanoma has the highest propensity of all cancers to metastasize to the brain with a large percentage of late-stage patients developing metastases in the central nervous system (CNS). It is well known that metastasis establishment, cell survival, and progression are affected by tumour-host cell interactions where changes in the host cellular compartments likely play an important role. In this context, miRNAs transferred by tumour derived extracellular vesicles (EVs) have previously been shown to create a favourable tumour microenvironment. Here, we show that miR-146a-5p is highly expressed in human melanoma brain metastasis (MBM) EVs, both in MBM cell lines as well as in biopsies, thereby modulating the brain metastatic niche. Mechanistically, miR-146a-5p was transferred to astrocytes via EV delivery and inhibited NUMB in the Notch signalling pathway. This resulted in activation of tumour-promoting cytokines (IL-6, IL-8, MCP-1 and CXCL1). Brain metastases were significantly reduced following miR-146a-5p knockdown. Corroborating these findings, miR-146a-5p inhibition led to a reduction of IL-6, IL-8, MCP-1 and CXCL1 in astrocytes. Following molecular docking analysis, deserpidine was identified as a functional miR-146a-5p inhibitor, both in vitro and in vivo. Our results highlight the pro-metastatic function of miR-146a-5p in EVs and identifies deserpidine for targeted adjuvant treatment.
Keywords: brain metastasis; deserpidine; extracellular vesicles; melanoma; miR-146a-5p; normal human astrocytes (NHA).
© 2023 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Achrol, A. S. , Rennert, R. C. , Anders, C. , Soffietti, R. , Ahluwalia, M. S. , Nayak, L. , Peters, S. , Arvold, N. D. , Harsh, G. R. , Steeg, P. S. , & Chang, S. D. (2019). Brain metastases. Nature Reviews Disease Primers, 5, 5. - PubMed
-
- Anvari, A. , Sasanpour, P. , & Rajabzadeh Kheradmardi, M. (2021). Radiotherapy and immunotherapy in melanoma brain metastases. Hematology/Oncology and Stem Cell Therapy, 16, 1–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

