Modeling of FAN1-Deficient Kidney Disease Using a Human Induced Pluripotent Stem Cell-Derived Kidney Organoid System
- PMID: 37759541
- PMCID: PMC10529520
- DOI: 10.3390/cells12182319
Modeling of FAN1-Deficient Kidney Disease Using a Human Induced Pluripotent Stem Cell-Derived Kidney Organoid System
Abstract
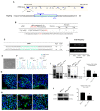
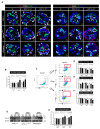
Karyomegalic interstitial nephritis (KIN) is a genetic kidney disease caused by mutations in the FANCD2/FANCI-Associated Nuclease 1 (FAN1) gene on 15q13.3, which results in karyomegaly and fibrosis of kidney cells through the incomplete repair of DNA damage. The aim of this study was to explore the possibility of using a human induced pluripotent stem cell (hiPSC)-derived kidney organoid system for modeling FAN1-deficient kidney disease, also known as KIN. We generated kidney organoids using WTC-11 (wild-type) hiPSCs and FAN1-mutant hiPSCs which include KIN patient-derived hiPSCs and FAN1-edited hiPSCs (WTC-11 FAN1+/-), created using the CRISPR/Cas9 system in WTC-11-hiPSCs. Kidney organoids from each group were treated with 20 nM of mitomycin C (MMC) for 24 or 48 h, and the expression levels of Ki67 and H2A histone family member X (H2A.X) were analyzed to detect DNA damage and assess the viability of cells within the kidney organoids. Both WTC-11-hiPSCs and FAN1-mutant hiPSCs were successfully differentiated into kidney organoids without structural deformities. MMC treatment for 48 h significantly increased the expression of DNA damage markers, while cell viability in both FAN1-mutant kidney organoids was decreased. However, these findings were observed in WTC-11-kidney organoids. These results suggest that FAN1-mutant kidney organoids can recapitulate the phenotype of FAN1-deficient kidney disease.
Keywords: FAN1 gene; induced pluripotent stem cells; karyomegalic interstitial nephritis; kidney organoid.
Conflict of interest statement
All the authors including Lee K.S. and Lee J.Y. of ToolGen company declare no competing financial interests or personal relationships that could influence this research.
Figures


Similar articles
-
Karyomegalic interstitial nephritis and DNA damage-induced polyploidy in Fan1 nuclease-defective knock-in mice.Genes Dev. 2016 Mar 15;30(6):639-44. doi: 10.1101/gad.276287.115. Genes Dev. 2016. PMID: 26980188 Free PMC article.
-
Generation of a human induced pluripotent stem cell line (CMCi001-A) from a patient with karyomegalic interstitial nephritis with homozygous frameshift deletion mutation c.1985_1994del10 of the FANCD2/FANCI-Associated Nuclease 1 gene.Stem Cell Res. 2020 Jul;46:101876. doi: 10.1016/j.scr.2020.101876. Epub 2020 Jun 12. Stem Cell Res. 2020. PMID: 32563974
-
New familial cases of karyomegalic interstitial nephritis with mutations in the FAN1 gene.BMC Med Genomics. 2021 Jun 14;14(1):160. doi: 10.1186/s12920-021-01009-7. BMC Med Genomics. 2021. PMID: 34126972 Free PMC article.
-
FAN1, a DNA Repair Nuclease, as a Modifier of Repeat Expansion Disorders.J Huntingtons Dis. 2021;10(1):95-122. doi: 10.3233/JHD-200448. J Huntingtons Dis. 2021. PMID: 33579867 Free PMC article. Review.
-
IgA Nephropathy Concomitant With Karyomegalic Interstitial Nephritis.Am J Med Sci. 2020 Sep;360(3):287-292. doi: 10.1016/j.amjms.2020.04.010. Epub 2020 Apr 17. Am J Med Sci. 2020. PMID: 32387117 Review.
Cited by
-
Trends and challenges in organoid modeling and expansion with pluripotent stem cells and somatic tissue.PeerJ. 2024 Nov 27;12:e18422. doi: 10.7717/peerj.18422. eCollection 2024. PeerJ. 2024. PMID: 39619184 Free PMC article. Review.
-
Kidney organoids: steps towards better organization and function.Biochem Soc Trans. 2024 Aug 28;52(4):1861-1871. doi: 10.1042/BST20231554. Biochem Soc Trans. 2024. PMID: 38934505 Free PMC article. Review.
-
Impact of exercise on immune cell infiltration in muscle tissue: implications for muscle repair and chronic disease.Clin Exp Med. 2025 Aug 27;25(1):306. doi: 10.1007/s10238-025-01852-3. Clin Exp Med. 2025. PMID: 40864296 Free PMC article. Review.
-
Organoids in Genetic Disorders: from Disease Modeling to Translational Applications.Stem Cell Rev Rep. 2025 Sep 11. doi: 10.1007/s12015-025-10973-x. Online ahead of print. Stem Cell Rev Rep. 2025. PMID: 40931310 Review.
-
Progress of Induced Pluripotent Stem Cell-Derived Renal Organoids in Clinical Application.Kidney Dis (Basel). 2024 Nov 11;11(1):1-10. doi: 10.1159/000541919. eCollection 2025 Jan-Dec. Kidney Dis (Basel). 2024. PMID: 40093027 Free PMC article. Review.
References
-
- Zhou W., Otto E.A., Cluckey A., Airik R., Hurd T.W., Chaki M., Diaz K., Lach F.P., Bennett G.R., Gee H.Y., et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 2012;44:910–915. doi: 10.1038/ng.2347. - DOI - PMC - PubMed
-
- Thongthip S., Bellani M., Gregg S.Q., Sridhar S., Conti B.A., Chen Y., Seidman M.M., Smogorzewska A. Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 2016;30:645–659. doi: 10.1101/gad.276261.115. - DOI - PMC - PubMed
-
- Kratz K., Schopf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavo E., Sartori A.A., Hengartner M.O., Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

