Evaluation of Linear Programming and Optimal Contribution Selection Approaches for Long-Term Selection on Beef Cattle Breeding
- PMID: 37759557
- PMCID: PMC10525978
- DOI: 10.3390/biology12091157
Evaluation of Linear Programming and Optimal Contribution Selection Approaches for Long-Term Selection on Beef Cattle Breeding
Abstract
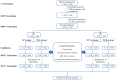
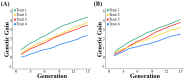
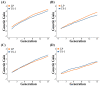
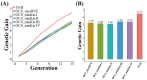
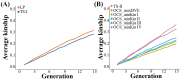
The optimized selection method can maximize the genetic gain in offspring under the premise of controlling the inbreeding level of the population. At present, genetic gain has been largely improved by using genomic selection in multiple farm animals. However, the design of the optimal selection method and assessment of its effects during long-term selection in beef cattle breeding are yet to be fully explored. In this study, a simulated beef cattle population was constructed, and 15 generations of simulated breeding were carried out using the linear programming breeding strategy (LP) and optimal contribution selection strategy (OCS), respectively. The truncation selection strategy (TS-I and TS-II) was used as the control. During the breeding process, genetic parameters including genetic gain, average kinship coefficient, QTL effect variance, and average observed heterozygosity were calculated and compared across generations. Our results showed that the LP method can significantly improve the genetic gain in the population, especially the genetic performance of the traits with high heritability and the traits with high weight in the breeding process, but the inbreeding level of the population is higher under LP strategy. Although the genetic gain in the population under the OCS strategy is lower than the TS-II strategy, this method can effectively control the inbreeding level of the population. Our findings also suggest that the LP and OCS method can be used as an effective means to improve genetic gain, while the OCS method is a more ideal method to obtain sustainable genetic gain during long-term selection.
Keywords: average kinship coefficient; cattle breeding; genetic gain; linear programming; optimal contribution selection; simulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Garcia-Ruiz A., Cole J.B., VanRaden P.M., Wiggans G.R., Ruiz-Lopez F.J., Van Tassell C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA. 2016;113:E4928. doi: 10.1073/pnas.1519061113. - DOI - PMC - PubMed
-
- Samore A.B., Fontanesi L. Genomic selection in pigs: State of the art and perspectives. Ital. J. Anim. Sci. 2016;15:211–232. doi: 10.1080/1828051X.2016.1172034. - DOI
-
- Daetwyler H.D., Hickey J.M., Henshall J.M., Dominik S., Gredler B., van der Werf J.H.J., Hayes B.J. Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Anim. Prod. Sci. 2010;50:1004–1010. doi: 10.1071/AN10096. - DOI
LinkOut - more resources
Full Text Sources

