Efforts to Minimise the Bacterial Genome as a Free-Living Growing System
- PMID: 37759570
- PMCID: PMC10525146
- DOI: 10.3390/biology12091170
Efforts to Minimise the Bacterial Genome as a Free-Living Growing System
Abstract
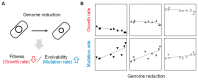
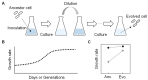
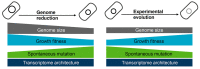
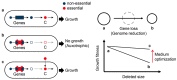
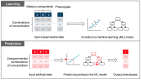
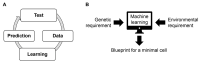
Exploring the minimal genetic requirements for cells to maintain free living is an exciting topic in biology. Multiple approaches are employed to address the question of the minimal genome. In addition to constructing the synthetic genome in the test tube, reducing the size of the wild-type genome is a practical approach for obtaining the essential genomic sequence for living cells. The well-studied Escherichia coli has been used as a model organism for genome reduction owing to its fast growth and easy manipulation. Extensive studies have reported how to reduce the bacterial genome and the collections of genomic disturbed strains acquired, which were sufficiently reviewed previously. However, the common issue of growth decrease caused by genetic disturbance remains largely unaddressed. This mini-review discusses the considerable efforts made to improve growth fitness, which was decreased due to genome reduction. The proposal and perspective are clarified for further accumulated genetic deletion to minimise the Escherichia coli genome in terms of genome reduction, experimental evolution, medium optimization, and machine learning.
Keywords: culture medium; experimental evolution; genome reduction; growth fitness; machine learning; minimal genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Experimental Challenges for Reduced Genomes: The Cell Model Escherichia coli.Microorganisms. 2019 Dec 18;8(1):3. doi: 10.3390/microorganisms8010003. Microorganisms. 2019. PMID: 31861355 Free PMC article. Review.
-
Global coordination of the mutation and growth rates across the genetic and nutritional variety in Escherichia coli.Front Microbiol. 2022 Sep 20;13:990969. doi: 10.3389/fmicb.2022.990969. eCollection 2022. Front Microbiol. 2022. PMID: 36204613 Free PMC article.
-
Correlation between genome reduction and bacterial growth.DNA Res. 2016 Dec;23(6):517-525. doi: 10.1093/dnares/dsw035. Epub 2016 Jul 3. DNA Res. 2016. PMID: 27374613 Free PMC article.
-
Coordinated Changes in Mutation and Growth Rates Induced by Genome Reduction.mBio. 2017 Jul 5;8(4):e00676-17. doi: 10.1128/mBio.00676-17. mBio. 2017. PMID: 28679744 Free PMC article.
-
Minimal genome: Worthwhile or worthless efforts toward being smaller?Biotechnol J. 2016 Feb;11(2):199-211. doi: 10.1002/biot.201400838. Epub 2015 Sep 10. Biotechnol J. 2016. PMID: 26356135 Review.
Cited by
-
Experimental evolution for the recovery of growth loss due to genome reduction.Elife. 2024 May 1;13:RP93520. doi: 10.7554/eLife.93520. Elife. 2024. PMID: 38690805 Free PMC article.
References
-
- Campillo-Balderas J.A., Lazcano A., Becerra A. Viral genome size distribution does not correlate with the antiquity of the host lineages. Front. Ecol. Evol. 2015;3:143. doi: 10.3389/fevo.2015.00143. - DOI
-
- Pellicer J., Fay M.F., Leitch I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010;164:10–15. doi: 10.1111/j.1095-8339.2010.01072.x. - DOI
-
- Rodríguez-Gijón A., Nuy J.K., Mehrshad M., Buck M., Schulz F., Woyke T., Garcia S.L. A Genomic Perspective Across Earth’s Microbiomes Reveals That Genome Size in Archaea and Bacteria Is Linked to Ecosystem Type and Trophic Strategy. Front. Microbiol. 2021;12:761869. doi: 10.3389/fmicb.2021.761869. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

