Integration of Omics Data and Network Models to Unveil Negative Aspects of SARS-CoV-2, from Pathogenic Mechanisms to Drug Repurposing
- PMID: 37759595
- PMCID: PMC10525644
- DOI: 10.3390/biology12091196
Integration of Omics Data and Network Models to Unveil Negative Aspects of SARS-CoV-2, from Pathogenic Mechanisms to Drug Repurposing
Abstract
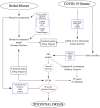
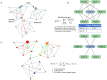
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the COVID-19 health emergency, affecting and killing millions of people worldwide. Following SARS-CoV-2 infection, COVID-19 patients show a spectrum of symptoms ranging from asymptomatic to very severe manifestations. In particular, bronchial and pulmonary cells, involved at the initial stage, trigger a hyper-inflammation phase, damaging a wide range of organs, including the heart, brain, liver, intestine and kidney. Due to the urgent need for solutions to limit the virus' spread, most efforts were initially devoted to mapping outbreak trajectories and variant emergence, as well as to the rapid search for effective therapeutic strategies. Samples collected from hospitalized or dead COVID-19 patients from the early stages of pandemic have been analyzed over time, and to date they still represent an invaluable source of information to shed light on the molecular mechanisms underlying the organ/tissue damage, the knowledge of which could offer new opportunities for diagnostics and therapeutic designs. For these purposes, in combination with clinical data, omics profiles and network models play a key role providing a holistic view of the pathways, processes and functions most affected by viral infection. In fact, in addition to epidemiological purposes, networks are being increasingly adopted for the integration of multiomics data, and recently their use has expanded to the identification of drug targets or the repositioning of existing drugs. These topics will be covered here by exploring the landscape of SARS-CoV-2 survey-based studies using systems biology approaches derived from omics data, paying particular attention to those that have considered samples of human origin.
Keywords: COVID-19; SARS-CoV-2; drug repurposing; networks; omics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Deciphering molecular mechanisms of SARS-CoV-2 pathogenesis and drug repurposing through GRN motifs: a comprehensive systems biology study.3 Biotech. 2023 Apr;13(4):117. doi: 10.1007/s13205-023-03518-x. Epub 2023 Mar 13. 3 Biotech. 2023. PMID: 37070032 Free PMC article.
-
Multiomics integration-based molecular characterizations of COVID-19.Brief Bioinform. 2022 Jan 17;23(1):bbab485. doi: 10.1093/bib/bbab485. Brief Bioinform. 2022. PMID: 34864875 Free PMC article. Review.
-
Cyclosporine A Inhibits Viral Infection and Release as Well as Cytokine Production in Lung Cells by Three SARS-CoV-2 Variants.Microbiol Spectr. 2022 Feb 23;10(1):e0150421. doi: 10.1128/spectrum.01504-21. Epub 2022 Jan 5. Microbiol Spectr. 2022. PMID: 34985303 Free PMC article.
-
Molecular Insights of SARS-CoV-2 Infection and Molecular Treatments.Curr Mol Med. 2022;22(7):621-639. doi: 10.2174/1566524021666211013121831. Curr Mol Med. 2022. PMID: 34645374 Review.
Cited by
-
An In-Depth Insight into Clinical, Cellular and Molecular Factors in COVID19-Associated Cardiovascular Ailments for Identifying Novel Disease Biomarkers, Drug Targets and Clinical Management Strategies.Arch Microbiol Immunol. 2024;8(3):290-308. doi: 10.26502/ami.936500177. Epub 2024 Jul 19. Arch Microbiol Immunol. 2024. PMID: 39703820 Free PMC article.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

