ABCD1 Transporter Deficiency Results in Altered Cholesterol Homeostasis
- PMID: 37759733
- PMCID: PMC10526550
- DOI: 10.3390/biom13091333
ABCD1 Transporter Deficiency Results in Altered Cholesterol Homeostasis
Abstract
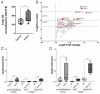
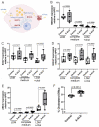
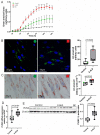
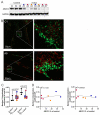
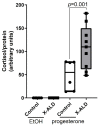
X-linked adrenoleukodystrophy (X-ALD), the most common peroxisomal disorder, is caused by mutations in the peroxisomal transporter ABCD1, resulting in the accumulation of very long-chain fatty acids (VLCFA). Strongly affected cell types, such as oligodendrocytes, adrenocortical cells and macrophages, exhibit high cholesterol turnover. Here, we investigated how ABCD1 deficiency affects cholesterol metabolism in human X-ALD patient-derived fibroblasts and CNS tissues of Abcd1-deficient mice. Lipidome analyses revealed increased levels of cholesterol esters (CE), containing both saturated VLCFA and mono/polyunsaturated (V)LCFA. The elevated CE(26:0) and CE(26:1) levels remained unchanged in LXR agonist-treated Abcd1 KO mice despite reduced total C26:0. Under high-cholesterol loading, gene expression of SOAT1, converting cholesterol to CE and lipid droplet formation were increased in human X-ALD fibroblasts versus healthy control fibroblasts. However, the expression of NCEH1, catalysing CE hydrolysis and the cholesterol transporter ABCA1 and cholesterol efflux were also upregulated. Elevated Soat1 and Abca1 expression and lipid droplet content were confirmed in the spinal cord of X-ALD mice, where expression of the CNS cholesterol transporter Apoe was also elevated. The extent of peroxisome-lipid droplet co-localisation appeared low and was not impaired by ABCD1-deficiency in cholesterol-loaded primary fibroblasts. Finally, addressing steroidogenesis, progesterone-induced cortisol release was amplified in X-ALD fibroblasts. These results link VLCFA to cholesterol homeostasis and justify further consideration of therapeutic approaches towards reducing VLCFA and cholesterol levels in X-ALD.
Keywords: ABCD1; Abcd1 KO mice; VLCFA; X-linked adrenoleukodystrophy; cholesterol esters; cortisol; lipid droplets; lipid metabolism; lipidomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kemp S., Pujol A., Waterham H.R., Van Geel B.M., Boehm C.D., Raymond G.V., Cutting G.R., Wanders R.J.A., Moser H.W. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: Role in diagnosis and clinical correlations. Hum. Mutat. 2001;18:499–515. doi: 10.1002/humu.1227. - DOI - PubMed
-
- Wiesinger C., Kunze M., Regelsberger G., Forss-Petter S., Berger J. Impaired very long-chain acyl-CoA β-oxidation in human X-linked adrenoleukodystrophy fibroblasts is a direct consequence of ABCD1 transporter dysfunction. J. Biol. Chem. 2013;288:19269–19279. doi: 10.1074/jbc.M112.445445. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

