Comparison, Analysis, and Molecular Dynamics Simulations of Structures of a Viral Protein Modeled Using Various Computational Tools
- PMID: 37760106
- PMCID: PMC10525864
- DOI: 10.3390/bioengineering10091004
Comparison, Analysis, and Molecular Dynamics Simulations of Structures of a Viral Protein Modeled Using Various Computational Tools
Abstract
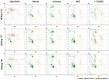
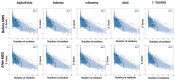
The structural analysis of proteins is a major domain of biomedical research. Such analysis requires resolved three-dimensional structures of proteins. Advancements in computer technology have led to progress in biomedical research. In silico prediction and modeling approaches have facilitated the construction of protein structures, with or without structural templates. In this study, we used three neural network-based de novo modeling approaches-AlphaFold2 (AF2), Robetta-RoseTTAFold (Robetta), and transform-restrained Rosetta (trRosetta)-and two template-based tools-the Molecular Operating Environment (MOE) and iterative threading assembly refinement (I-TASSER)-to construct the structure of a viral capsid protein, hepatitis C virus core protein (HCVcp), whose structure have not been fully resolved by laboratory techniques. Templates with sufficient sequence identity for the homology modeling of complete HCVcp are currently unavailable. Therefore, we performed domain-based homology modeling for MOE simulations. The templates for each domain were obtained through sequence-based searches on NCBI and the Protein Data Bank. Then, the modeled domains were assembled to construct the complete structure of HCVcp. The full-length structure and two truncated forms modeled using various computational tools were compared. Molecular dynamics (MD) simulations were performed to refine the structures. The root mean square deviation of backbone atoms, root mean square fluctuation of Cα atoms, and radius of gyration were calculated to monitor structural changes and convergence in the simulations. The model quality was evaluated through ERRAT and phi-psi plot analysis. In terms of the initial prediction for protein modeling, Robetta and trRosetta outperformed AF2. Regarding template-based tools, MOE outperformed I-TASSER. MD simulations resulted in compactly folded protein structures, which were of good quality and theoretically accurate. Thus, the predicted structures of certain proteins must be refined to obtain reliable structural models. MD simulation is a promising tool for this purpose.
Keywords: AF2; HCV core protein; I-TASSER; MD simulation; MOE; Robetta-RoseTTAFold; homology modeling; trRosetta.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources

