Transcriptome Studies Reveal the N6-Methyladenosine Differences in Testis of Yaks at Juvenile and Sexual Maturity Stages
- PMID: 37760215
- PMCID: PMC10525320
- DOI: 10.3390/ani13182815
Transcriptome Studies Reveal the N6-Methyladenosine Differences in Testis of Yaks at Juvenile and Sexual Maturity Stages
Abstract
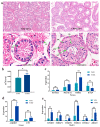
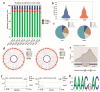
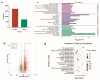
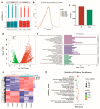
Studying the mechanism of spermatogenesis is key to exploring the reproductive characteristics of male yaks. Although N6-methyladenosine (m6A) RNA modification has been reported to regulate spermatogenesis and reproductive function in mammals, the molecular mechanism of m6A in yak testis development and spermatogenesis remains largely unknown. Therefore, we collected testicular tissue from juvenile and adult yaks and found that the m6A level significantly increased after sexual maturity in yaks. In MeRIP-seq, 1702 hypermethylated peaks and 724 hypomethylated peaks were identified. The hypermethylated differentially methylated RNAs (DMRs) (CIB2, AK1, FOXJ2, PKDREJ, SLC9A3, and TOPAZ1) mainly regulated spermatogenesis. Functional enrichment analysis showed that DMRs were significantly enriched in the adherens junction, gap junction, and Wnt, PI3K, and mTOR signaling pathways, regulating cell development, spermatogenesis, and testicular endocrine function. The functional analysis of differentially expressed genes showed that they were involved in the biological processes of mitosis, meiosis, and flagellated sperm motility during the sexual maturity of yak testis. We also screened the key regulatory factors of testis development and spermatogenesis by combined analysis, which included BRCA1, CREBBP, STAT3, and SMAD4. This study indexed the m6A characteristics of yak testicles at different developmental stages, providing basic data for further research of m6A modification regulating yak testicular development.
Keywords: N6-methyladenosine; mRNA; reproduction; spermatogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Characterization of N6-methyladenosine in cattle-yak testis tissue.Front Vet Sci. 2022 Aug 9;9:971515. doi: 10.3389/fvets.2022.971515. eCollection 2022. Front Vet Sci. 2022. PMID: 36016801 Free PMC article.
-
Characterization of N6-Methyladenosine in Domesticated Yak Testes Before and After Sexual Maturity.Front Cell Dev Biol. 2021 Nov 11;9:755670. doi: 10.3389/fcell.2021.755670. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34858983 Free PMC article.
-
Comparison of reproductive performance and functional analysis of spermatogenesis factors between domestic yak and semi-wild blood yak.BMC Genomics. 2025 Apr 29;26(1):418. doi: 10.1186/s12864-025-11594-x. BMC Genomics. 2025. PMID: 40301732 Free PMC article.
-
RNA N6-methyladenosine modification, spermatogenesis, and human male infertility.Mol Hum Reprod. 2021 May 29;27(6):gaab020. doi: 10.1093/molehr/gaab020. Mol Hum Reprod. 2021. PMID: 33749751 Review.
-
Connecting the Dots: N6-Methyladenosine (m6 A) Modification in Spermatogenesis.Adv Biol (Weinh). 2023 Aug;7(8):e2300068. doi: 10.1002/adbi.202300068. Epub 2023 Jun 23. Adv Biol (Weinh). 2023. PMID: 37353958 Review.
Cited by
-
Characteristic analysis of N6-methyladenine in different parts of yak epididymis.BMC Genomics. 2025 May 19;26(1):500. doi: 10.1186/s12864-025-11684-w. BMC Genomics. 2025. PMID: 40389816 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

