Glutamine Regulates Gene Expression Profiles to Increase the Proliferation of Porcine Intestinal Epithelial Cells and the Expansion of Intestinal Stem Cells
- PMID: 37760316
- PMCID: PMC10525449
- DOI: 10.3390/ani13182917
Glutamine Regulates Gene Expression Profiles to Increase the Proliferation of Porcine Intestinal Epithelial Cells and the Expansion of Intestinal Stem Cells
Abstract
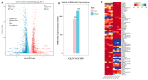
The intestinal epithelium is known for its rapid self-renewal, and glutamine is crucial in providing carbon and nitrogen for biosynthesis. However, understanding how glutamine affects gene expression in the intestinal epithelium is limited, and identifying the essential genes and signals involved in regulating intestinal epithelial cell growth is particularly challenging. In this study, glutamine supplementation exhibited a robust acceleration of intestinal epithelial cell proliferation and stem cell expansion. RNA sequencing indicated diverse transcriptome changes between the control and glutamine supplementation groups, identifying 925 up-regulated and 1152 down-regulated genes. The up-regulated DEGs were enriched in the KEGG pathway of cell cycle and GO terms of DNA replication initiation, regulation of phosphatidylinositol 3-kinase activity, DNA replication, minichromosome maintenance protein (MCM) complex, and ATP binding, whereas the down-regulated DEGs were enriched in the KEGG pathway of p53 signaling pathway, TNF signaling pathway, and JAK-STAT signaling pathway and GO terms of inflammatory response and intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress. Furthermore, GSEA analysis revealed a significant up-regulation of the cell cycle, DNA replication initiation, ATP-dependent RNA helicase activity, and down-regulation of the TNF signaling pathway. The protein-protein association network of the intersecting genes highlighted the significance of DNA replication licensing factors (MCM3, MCM6, and MCM10) in promoting intestinal epithelial growth in response to glutamine. Based on these findings, we propose that glutamine may upregulate DNA replication licensing factors, leading to increased PI3K/Akt signaling and the suppression of TNF, JAK-STAT, and p53 pathways. Consequently, this mechanism results in the proliferation of porcine intestinal epithelial cells and the expansion of intestinal stem cells.
Keywords: gene expression profile; glutamine; intestinal epithelium; intestinal stem cells; minichromosome maintenance protein; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Pentinmikko N., Iqbal S., Mana M., Andersson S., Cognetta A.B., 3rd, Suciu R.M., Roper J., Luopajärvi K., Markelin E., Gopalakrishnan S., et al. Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature. 2019;571:398–402. doi: 10.1038/s41586-019-1383-0. - DOI - PMC - PubMed
Grants and funding
- 32202710; 32002213/National Natural Science Foundation of China
- Qiankehe foundation-ZK [2022] General 159/Science and Technology Project of Guizhou Province
- Gui Da Te Gang He Zi [2020] 24/Natural Science Special Research Fund of Guizhou University
- Qiannong Finance (2022) No. 10/Guizhou Provincial Finance Seed Industry Development Project
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

