PorcineAI-Enhancer: Prediction of Pig Enhancer Sequences Using Convolutional Neural Networks
- PMID: 37760334
- PMCID: PMC10526013
- DOI: 10.3390/ani13182935
PorcineAI-Enhancer: Prediction of Pig Enhancer Sequences Using Convolutional Neural Networks
Abstract
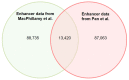
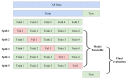
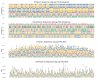
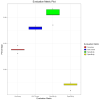
Understanding the mechanisms of gene expression regulation is crucial in animal breeding. Cis-regulatory DNA sequences, such as enhancers, play a key role in regulating gene expression. Identifying enhancers is challenging, despite the use of experimental techniques and computational methods. Enhancer prediction in the pig genome is particularly significant due to the costliness of high-throughput experimental techniques. The study constructed a high-quality database of pig enhancers by integrating information from multiple sources. A deep learning prediction framework called PorcineAI-enhancer was developed for the prediction of pig enhancers. This framework employs convolutional neural networks for feature extraction and classification. PorcineAI-enhancer showed excellent performance in predicting pig enhancers, validated on an independent test dataset. The model demonstrated reliable prediction capability for unknown enhancer sequences and performed remarkably well on tissue-specific enhancer sequences.The study developed a deep learning prediction framework, PorcineAI-enhancer, for predicting pig enhancers. The model demonstrated significant predictive performance and potential for tissue-specific enhancers. This research provides valuable resources for future studies on gene expression regulation in pigs.
Keywords: convolutional neural networks; enhancer; sequence classification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ADH-Enhancer: an attention-based deep hybrid framework for enhancer identification and strength prediction.Brief Bioinform. 2024 Jan 22;25(2):bbae030. doi: 10.1093/bib/bbae030. Brief Bioinform. 2024. PMID: 38385876 Free PMC article.
-
RicENN: Prediction of Rice Enhancers with Neural Network Based on DNA Sequences.Interdiscip Sci. 2022 Jun;14(2):555-565. doi: 10.1007/s12539-022-00503-5. Epub 2022 Feb 21. Interdiscip Sci. 2022. PMID: 35190950
-
A multi-perspective deep learning framework for enhancer characterization and identification.Comput Biol Chem. 2025 Feb;114:108284. doi: 10.1016/j.compbiolchem.2024.108284. Epub 2024 Nov 19. Comput Biol Chem. 2025. PMID: 39577030
-
Progress and challenges in bioinformatics approaches for enhancer identification.Brief Bioinform. 2016 Nov;17(6):967-979. doi: 10.1093/bib/bbv101. Epub 2015 Dec 3. Brief Bioinform. 2016. PMID: 26634919 Free PMC article. Review.
-
Sequence-Based Deep Learning Frameworks on Enhancer-Promoter Interactions Prediction.Curr Pharm Des. 2021;27(15):1847-1855. doi: 10.2174/1381612826666201124112710. Curr Pharm Des. 2021. PMID: 33234095 Review.
Cited by
-
From COVID-19 to monkeypox: a novel predictive model for emerging infectious diseases.BioData Min. 2024 Oct 22;17(1):42. doi: 10.1186/s13040-024-00396-8. BioData Min. 2024. PMID: 39438943 Free PMC article.
References
-
- Beagan J.A., Pastuzyn E.D., Fernandez L.R., Guo M.H., Feng K., Titus K.R., Chandrashekar H., Shepherd J.D., Phillips-Cremins J.E. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat. Neurosci. 2020;23:707–717. doi: 10.1038/s41593-020-0634-6. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

