Artificial Intelligence in Digital Pathology for Bladder Cancer: Hype or Hope? A Systematic Review
- PMID: 37760487
- PMCID: PMC10526515
- DOI: 10.3390/cancers15184518
Artificial Intelligence in Digital Pathology for Bladder Cancer: Hype or Hope? A Systematic Review
Abstract
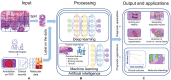
Bladder cancer (BC) diagnosis and prediction of prognosis are hindered by subjective pathological evaluation, which may cause misdiagnosis and under-/over-treatment. Computational pathology (CPATH) can identify clinical outcome predictors, offering an objective approach to improve prognosis. However, a systematic review of CPATH in BC literature is lacking. Therefore, we present a comprehensive overview of studies that used CPATH in BC, analyzing 33 out of 2285 identified studies. Most studies analyzed regions of interest to distinguish normal versus tumor tissue and identify tumor grade/stage and tissue types (e.g., urothelium, stroma, and muscle). The cell's nuclear area, shape irregularity, and roundness were the most promising markers to predict recurrence and survival based on selected regions of interest, with >80% accuracy. CPATH identified molecular subtypes by detecting features, e.g., papillary structures, hyperchromatic, and pleomorphic nuclei. Combining clinicopathological and image-derived features improved recurrence and survival prediction. However, due to the lack of outcome interpretability and independent test datasets, robustness and clinical applicability could not be ensured. The current literature demonstrates that CPATH holds the potential to improve BC diagnosis and prediction of prognosis. However, more robust, interpretable, accurate models and larger datasets-representative of clinical scenarios-are needed to address artificial intelligence's reliability, robustness, and black box challenge.
Keywords: artificial intelligence; bladder cancer; computational pathology; computer-aided diagnosis; digital pathology; histopathology; image analysis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Woerl A.-C., Eckstein M., Geiger J., Wagner D.C., Daher T., Stenzel P., Fernandez A., Hartmann A., Wand M., Roth W., et al. Deep Learning Predicts Molecular Subtype of Muscle-invasive Bladder Cancer from Conventional Histopathological Slides. Eur. Urol. 2020;78:256–264. doi: 10.1016/j.eururo.2020.04.023. - DOI - PubMed
-
- Zhang Z., Chen P., McGough M., Xing F., Wang C., Bui M., Xie Y., Sapkota M., Cui L., Dhillon J., et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 2019;1:236–245. doi: 10.1038/s42256-019-0052-1. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

