Repurposing Clemastine to Target Glioblastoma Cell Stemness
- PMID: 37760589
- PMCID: PMC10526458
- DOI: 10.3390/cancers15184619
Repurposing Clemastine to Target Glioblastoma Cell Stemness
Abstract
Brain tumor-initiating cells (BTICs) and tumor cell plasticity promote glioblastoma (GBM) progression. Here, we demonstrate that clemastine, an over-the-counter drug for treating hay fever and allergy symptoms, effectively attenuated the stemness and suppressed the propagation of primary BTIC cultures bearing PDGFRA amplification. These effects on BTICs were accompanied by altered gene expression profiling indicative of their more differentiated states, resonating with the activity of clemastine in promoting the differentiation of normal oligodendrocyte progenitor cells (OPCs) into mature oligodendrocytes. Functional assays for pharmacological targets of clemastine revealed that the Emopamil Binding Protein (EBP), an enzyme in the cholesterol biosynthesis pathway, is essential for BTIC propagation and a target that mediates the suppressive effects of clemastine. Finally, we showed that a neural stem cell-derived mouse glioma model displaying predominantly proneural features was similarly susceptible to clemastine treatment. Collectively, these results identify pathways essential for maintaining the stemness and progenitor features of GBMs, uncover BTIC dependency on EBP, and suggest that non-oncology, low-toxicity drugs with OPC differentiation-promoting activity can be repurposed to target GBM stemness and aid in their treatment.
Keywords: Emopamil Binding Protein (EBP); clemastine; glioblastoma; stemness.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
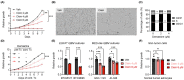

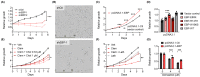
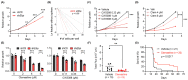
Similar articles
-
Clemastine Promotes Differentiation of Oligodendrocyte Progenitor Cells Through the Activation of ERK1/2 via Muscarinic Receptors After Spinal Cord Injury.Front Pharmacol. 2022 Jul 5;13:914153. doi: 10.3389/fphar.2022.914153. eCollection 2022. Front Pharmacol. 2022. PMID: 35865954 Free PMC article.
-
Comparative genomic and genetic analysis of glioblastoma-derived brain tumor-initiating cells and their parent tumors.Neuro Oncol. 2016 Mar;18(3):350-60. doi: 10.1093/neuonc/nov143. Epub 2015 Aug 5. Neuro Oncol. 2016. PMID: 26245525 Free PMC article.
-
Fibrinogen in the glioblastoma microenvironment contributes to the invasiveness of brain tumor-initiating cells.Brain Pathol. 2021 Sep;31(5):e12947. doi: 10.1111/bpa.12947. Epub 2021 Mar 10. Brain Pathol. 2021. PMID: 33694259 Free PMC article.
-
Novel concept of the border niche: glioblastoma cells use oligodendrocytes progenitor cells (GAOs) and microglia to acquire stem cell-like features.Brain Tumor Pathol. 2019 Apr;36(2):63-73. doi: 10.1007/s10014-019-00341-2. Epub 2019 Apr 9. Brain Tumor Pathol. 2019. PMID: 30968276 Review.
-
Oligodendrocyte Progenitor Cells in the Tumor Microenvironment.Adv Exp Med Biol. 2020;1234:107-122. doi: 10.1007/978-3-030-37184-5_8. Adv Exp Med Biol. 2020. PMID: 32040858 Review.
Cited by
-
A longitudinal single-cell and spatial multiomic atlas of pediatric high-grade glioma.bioRxiv [Preprint]. 2024 Mar 8:2024.03.06.583588. doi: 10.1101/2024.03.06.583588. bioRxiv. 2024. PMID: 38496580 Free PMC article. Preprint.
References
-
- Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

