Identification of Potentially Novel Molecular Targets of Endometrial Cancer Using a Non-Biased Proteomic Approach
- PMID: 37760635
- PMCID: PMC10527058
- DOI: 10.3390/cancers15184665
Identification of Potentially Novel Molecular Targets of Endometrial Cancer Using a Non-Biased Proteomic Approach
Abstract
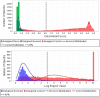
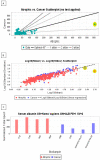
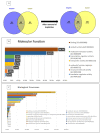
The present study was aimed at identifying novel proteins in endometrial cancer (EC), employing proteomic analysis of tissues obtained after surgery. A differential MS-based proteomic analysis was conducted from whole tissues dissected from biopsies from post-menopausal women, histologically confirmed as endometrial cancer (two endometrioid and two serous; n = 4) or normal atrophic endometrium (n = 4), providing 888 differentially expressed proteins with 246 of these previously documented elsewhere as expressed in EC and 372 proteins not previously demonstrated to be expressed in EC but associated with other types of cancer. Additionally, 33 proteins not recorded previously in PubMed as being expressed in any forms of cancer were also identified, with only 26 of these proteins having a publication associated with their expression patterns or putative functions. The putative functions of the 26 proteins (GRN, APP, HEXA, CST3, CAD, QARS, SIAE, WARS, MYH8, CLTB, GOLIM4, SCARB2, BOD1L1, C14orf142, C9orf142, CCDC13, CNPY4, FAM169A, HN1L, PIGT, PLCL1, PMFBP1, SARS2, SCPEP1, SLC25A24 and ZC3H4) in other tissues point towards and provide a basis for further investigation of these previously unrecognised novel EC proteins. The developmental biology, disease, extracellular matrix, homeostatic, immune, metabolic (both RNA and protein), programmed cell death, signal transduction, molecular transport, transcriptional networks and as yet uncharacterised pathways indicate that these proteins are potentially involved in endometrial carcinogenesis and thus may be important in EC diagnosis, prognostication and treatment and thus are worthy of further investigation.
Keywords: biomarker; endometrial cancer; prognosis; protein; proteomics; therapy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Proteomic analysis of stage I endometrial cancer tissue: identification of proteins associated with oxidative processes and inflammation.Gynecol Oncol. 2011 Jun 1;121(3):586-94. doi: 10.1016/j.ygyno.2011.02.031. Epub 2011 Apr 1. Gynecol Oncol. 2011. PMID: 21458040 Free PMC article.
-
The oncofetal protein IMP3: a novel biomarker for endometrial serous carcinoma.Am J Surg Pathol. 2008 Feb;32(2):304-15. doi: 10.1097/PAS.0b013e3181483ff8. Am J Surg Pathol. 2008. PMID: 18223334
-
Deciphering the proteomic signature of human endometrial receptivity.Hum Reprod. 2014 Sep;29(9):1957-67. doi: 10.1093/humrep/deu171. Epub 2014 Aug 8. Hum Reprod. 2014. PMID: 25106620
-
Novel molecular profiles of endometrial cancer-new light through old windows.J Steroid Biochem Mol Biol. 2008 Feb;108(3-5):221-9. doi: 10.1016/j.jsbmb.2007.09.020. Epub 2007 Sep 15. J Steroid Biochem Mol Biol. 2008. PMID: 18061438 Review.
-
The Role of KRAS in Endometrial Cancer: A Mini-Review.Anticancer Res. 2019 Feb;39(2):533-539. doi: 10.21873/anticanres.13145. Anticancer Res. 2019. PMID: 30711927 Review.
Cited by
-
Proteomic Profile of Endometrial Cancer: A Scoping Review.Biology (Basel). 2024 Aug 1;13(8):584. doi: 10.3390/biology13080584. Biology (Basel). 2024. PMID: 39194522 Free PMC article.
-
Shedding of GPP130 by PC7 and Furin: Potential Implication in Lung Cancer Progression.Int J Mol Sci. 2025 Jun 26;26(13):6164. doi: 10.3390/ijms26136164. Int J Mol Sci. 2025. PMID: 40649939 Free PMC article.
-
Expression, Distribution and Function of the Transient Receptor Potential Vanilloid Type 1 (TRPV1) in Endometrial Cancer.Int J Mol Sci. 2025 Mar 27;26(7):3104. doi: 10.3390/ijms26073104. Int J Mol Sci. 2025. PMID: 40243844 Free PMC article.
-
SIM2, associated with clinicopathologic features, promotes the malignant biological behaviors of endometrial carcinoma cells.BMC Cancer. 2025 Apr 11;25(1):666. doi: 10.1186/s12885-025-14077-0. BMC Cancer. 2025. PMID: 40217155 Free PMC article.
References
-
- Ricceri F., Giraudo M.T., Fasanelli F., Milanese D., Sciannameo V., Fiorini L., Sacerdote C. Diet and endometrial cancer: A focus on the role of fruit and vegetable intake, Mediterranean diet and dietary inflammatory index in the endometrial cancer risk. BMC Cancer. 2017;17:757. doi: 10.1186/s12885-017-3754-y. - DOI - PMC - PubMed
-
- Clarke M.A., Long B.J., Del Mar Morillo A., Arbyn M., Bakkum-Gamez J.N., Wentzensen N. Association of endometrial cancer risk with postmenopausal bleeding in women: A systematic review and meta-analysis. JAMA Intern. Med. 2018;178:1210–1222. doi: 10.1001/jamainternmed.2018.2820. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

