Pseudouridine Synthase RsuA Confers a Survival Advantage to Bacteria under Streptomycin Stress
- PMID: 37760743
- PMCID: PMC10525438
- DOI: 10.3390/antibiotics12091447
Pseudouridine Synthase RsuA Confers a Survival Advantage to Bacteria under Streptomycin Stress
Abstract
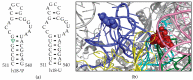
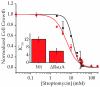
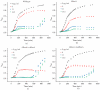
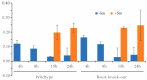
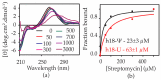
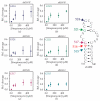
Bacterial ribosome small subunit rRNA (16S rRNA) contains 11 nucleotide modifications scattered throughout all its domains. The 16S rRNA pseudouridylation enzyme, RsuA, which modifies U516, is a survival protein essential for bacterial survival under stress conditions. A comparison of the growth curves of wildtype and RsuA knock-out E. coli strains illustrates that RsuA renders a survival advantage to bacteria under streptomycin stress. The RsuA-dependent growth advantage for bacteria was found to be dependent on its pseudouridylation activity. In addition, the role of RsuA as a trans-acting factor during ribosome biogenesis may also play a role in bacterial growth under streptomycin stress. Furthermore, circular dichroism spectroscopy measurements and RNase footprinting studies have demonstrated that pseudouridine at position 516 influences helix 18 structure, folding, and streptomycin binding. This study exemplifies the importance of bacterial rRNA modification enzymes during environmental stress.
Keywords: helix18; pseudouridine; pseudouridine synthase; ribosome; streptomycin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Douthwaite S., Fourmy D., Yoshizawa S. In: Fine-Tuning of RNA Functions by Modification and Editing. 2nd ed. Grosjean H., editor. Volume 12. Springer; Berlin/Heidelberg, Germany: 2005. pp. 285–307.
-
- Lázaro E., Rodriguez-Fonseca C., Porse B., Ureña D., Garrett R.A., Ballesta J.P.G. A Sparsomycin-resistant Mutant of Halobacterium salinarium Lacks a Modification at Nucleotide U2603 in the Peptidyl Transferase Centre of 23 S rRNA. J. Mol. Biol. 1996;261:231–238. doi: 10.1006/jmbi.1996.0455. - DOI - PubMed
LinkOut - more resources
Full Text Sources

