Discriminative Identification of SARS-CoV-2 Variants Based on Mass-Spectrometry Analysis
- PMID: 37760814
- PMCID: PMC10525290
- DOI: 10.3390/biomedicines11092373
Discriminative Identification of SARS-CoV-2 Variants Based on Mass-Spectrometry Analysis
Abstract
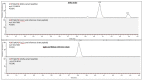
The spread of SARS-CoV-2 variants of concern (VOCs) is of great importance since genetic changes may increase transmissibility, disease severity and reduce vaccine effectiveness. Moreover, these changes may lead to failure of diagnostic measures. Therefore, variant-specific diagnostic methods are essential. To date, genetic sequencing is the gold-standard method to discriminate between variants. However, it is time-consuming (taking several days) and expensive. Therefore, the development of rapid diagnostic methods for SARS-CoV-2 in accordance with its genetic modification is of great importance. In this study we introduce a Mass Spectrometry (MS)-based methodology for the diagnosis of SARS-CoV-2 in propagated in cell-culture. This methodology enables the universal identification of SARS-CoV-2, as well as variant-specific discrimination. The universal identification of SARS-CoV-2 is based on conserved markers shared by all variants, while the identification of specific variants relies on variant-specific markers. Determining a specific set of peptides for a given variant consists of a multistep procedure, starting with an in-silico search for variant-specific tryptic peptides, followed by a tryptic digest of a cell-cultured SARS-CoV-2 variant, and identification of these markers by HR-LC-MS/MS analysis. As a proof of concept, this approach was demonstrated for four representative VOCs compared to the wild-type Wuhan reference strain. For each variant, at least two unique markers, derived mainly from the spike (S) and nucleocapsid (N) viral proteins, were identified. This methodology is specific, rapid, easy to perform and inexpensive. Therefore, it can be applied as a diagnostic tool for pathogenic variants.
Keywords: COVID19; LC-MS/MS; SARS-CoV-2; diagnosis; mass-spectrometry; nucleocapsid; spike; variants of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

