Establishing Molecular Subgroups of CD8+ T Cell-Associated Genes in the Ovarian Cancer Tumour Microenvironment and Predicting the Immunotherapy Response
- PMID: 37760841
- PMCID: PMC10525231
- DOI: 10.3390/biomedicines11092399
Establishing Molecular Subgroups of CD8+ T Cell-Associated Genes in the Ovarian Cancer Tumour Microenvironment and Predicting the Immunotherapy Response
Abstract
Background: The mechanism by which infiltrating CD8+ T lymphocytes in the tumour microenvironment influence the survival of patients with ovarian cancer (OC) remains unclear.
Methods: To identify biomarkers to optimise OC treatment, 13 immune-cell-line-associated datasets, RNA sequencing data, and clinical data from the GEO, TCGA, and the ICGC were collected. Gene expression in OC was assessed using quantitative reverse transcription polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC) staining.
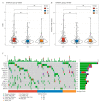
Results: We identified 520 genes and three immunological clusters (IC1, IC2, and IC3) associated with CD8+ T cells. Higher IFN scores, immune T cell lytic activity, and immune cell infiltration and upregulated expression of immune-checkpoint-related genes indicated that IC3 is more responsive to immunotherapy, whereas IC1 and IC2 have a poorer prognosis. A 10-gene signature, including SEMA4F, CX3CR1, STX7, PASK, AKIRIN2, HEMGN, GBP5, NSG1, and CXorf65, was constructed, and a multivariate Cox regression analysis revealed a significant association between the 10-gene signature-based risk model and overall survival (p < 0.001). A nomogram was constructed with age and the 10-gene signature. Consistent with the bioinformatics analysis, IHC and qRT-PCR confirmed the accuracy of the signatures in OC tissue samples. The predictive ability of the risk model was demonstrated using the Imvigor210 immunotherapy dataset.
Conclusions: The development of a novel gene signature associated with CD8+ T cells could facilitate more accurate prognostics and prediction of the immunotherapeutic response of patients with OC.
Keywords: biomarkers; immunotherapy; ovarian cancer; risk model; tumour microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Konstantinopoulos P.A., Ceccaldi R., Shapiro G.I., D’Andrea A.D. Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

