DSMRI: Domain Shift Analyzer for Multi-Center MRI Datasets
- PMID: 37761314
- PMCID: PMC10527875
- DOI: 10.3390/diagnostics13182947
DSMRI: Domain Shift Analyzer for Multi-Center MRI Datasets
Abstract
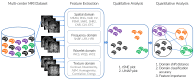
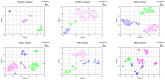
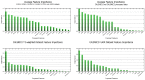
In medical research and clinical applications, the utilization of MRI datasets from multiple centers has become increasingly prevalent. However, inherent variability between these centers presents challenges due to domain shift, which can impact the quality and reliability of the analysis. Regrettably, the absence of adequate tools for domain shift analysis hinders the development and validation of domain adaptation and harmonization techniques. To address this issue, this paper presents a novel Domain Shift analyzer for MRI (DSMRI) framework designed explicitly for domain shift analysis in multi-center MRI datasets. The proposed model assesses the degree of domain shift within an MRI dataset by leveraging various MRI-quality-related metrics derived from the spatial domain. DSMRI also incorporates features from the frequency domain to capture low- and high-frequency information about the image. It further includes the wavelet domain features by effectively measuring the sparsity and energy present in the wavelet coefficients. Furthermore, DSMRI introduces several texture features, thereby enhancing the robustness of the domain shift analysis process. The proposed framework includes visualization techniques such as t-SNE and UMAP to demonstrate that similar data are grouped closely while dissimilar data are in separate clusters. Additionally, quantitative analysis is used to measure the domain shift distance, domain classification accuracy, and the ranking of significant features. The effectiveness of the proposed approach is demonstrated using experimental evaluations on seven large-scale multi-site neuroimaging datasets.
Keywords: MRI; UMAP; domain shift; quality control; t-SNE; texture analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Visual MRI: merging information visualization and non-parametric clustering techniques for MRI dataset analysis.Artif Intell Med. 2008 Nov;44(3):183-99. doi: 10.1016/j.artmed.2008.06.006. Epub 2008 Sep 4. Artif Intell Med. 2008. PMID: 18775655
-
Abbreviated Versus Multiparametric Prostate MRI in Active Surveillance for Prostate-Cancer Patients: Comparison of Accuracy and Clinical Utility as a Decisional Tool.Diagnostics (Basel). 2023 Feb 4;13(4):578. doi: 10.3390/diagnostics13040578. Diagnostics (Basel). 2023. PMID: 36832066 Free PMC article.
-
S-CUDA: Self-cleansing unsupervised domain adaptation for medical image segmentation.Med Image Anal. 2021 Dec;74:102214. doi: 10.1016/j.media.2021.102214. Epub 2021 Aug 12. Med Image Anal. 2021. PMID: 34464837
-
Deep Learning in Large and Multi-Site Structural Brain MR Imaging Datasets.Front Neuroinform. 2022 Jan 20;15:805669. doi: 10.3389/fninf.2021.805669. eCollection 2021. Front Neuroinform. 2022. PMID: 35126080 Free PMC article. Review.
-
From Silos to Synthesis: A comprehensive review of domain adaptation strategies for multi-source data integration in healthcare.Comput Biol Med. 2025 Jun;191:110108. doi: 10.1016/j.compbiomed.2025.110108. Epub 2025 Apr 9. Comput Biol Med. 2025. PMID: 40209575 Review.
Cited by
-
Toward diffusion tensor imaging as a biomarker in neurodegenerative diseases: technical considerations to optimize recordings and data processing.Front Hum Neurosci. 2024 Apr 2;18:1378896. doi: 10.3389/fnhum.2024.1378896. eCollection 2024. Front Hum Neurosci. 2024. PMID: 38628970 Free PMC article. Review.
References
-
- Kushol R., Masoumzadeh A., Huo D., Kalra S., Yang Y.H. Addformer: Alzheimer’s disease detection from structural Mri using fusion transformer; Proceedings of the IEEE 19th International Symposium on Biomedical Imaging; Kolkata, India. 28–31 March 2022; pp. 1–5.
-
- Kushol R., Luk C.C., Dey A., Benatar M., Briemberg H., Dionne A., Dupré N., Frayne R., Genge A., Gibson S., et al. SF2Former: Amyotrophic lateral sclerosis identification from multi-center MRI data using spatial and frequency fusion transformer. Comput. Med Imaging Graph. 2023;108:102279. doi: 10.1016/j.compmedimag.2023.102279. - DOI - PubMed
-
- Quinonero-Candela J., Sugiyama M., Schwaighofer A., Lawrence N.D. Dataset Shift in Machine Learning. MIT Press; Cambridge, MA, USA: 2008.
LinkOut - more resources
Full Text Sources

