Origins of Genetic Coding: Self-Guided Molecular Self-Organisation
- PMID: 37761580
- PMCID: PMC10527755
- DOI: 10.3390/e25091281
Origins of Genetic Coding: Self-Guided Molecular Self-Organisation
Abstract
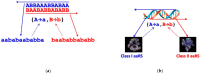
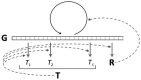
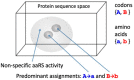
The origin of genetic coding is characterised as an event of cosmic significance in which quantum mechanical causation was transcended by constructive computation. Computational causation entered the physico-chemical processes of the pre-biotic world by the incidental satisfaction of a condition of reflexivity between polymer sequence information and system elements able to facilitate their own production through translation of that information. This event, which has previously been modelled in the dynamics of Gene-Replication-Translation systems, is properly described as a process of self-guided self-organisation. The spontaneous emergence of a primordial genetic code between two-letter alphabets of nucleotide triplets and amino acids is easily possible, starting with random peptide synthesis that is RNA-sequence-dependent. The evident self-organising mechanism is the simultaneous quasi-species bifurcation of the populations of information-carrying genes and enzymes with aminoacyl-tRNA synthetase-like activities. This mechanism allowed the code to evolve very rapidly to the ~20 amino acid limit apparent for the reflexive differentiation of amino acid properties using protein catalysts. The self-organisation of semantics in this domain of physical chemistry conferred on emergent molecular biology exquisite computational control over the nanoscopic events needed for its self-construction.
Keywords: aminoacyl-tRNA synthetase; computation; genetic coding; mechanistic causation; reflexivity; replication; translation.
Conflict of interest statement
The author declare no conflict of interest.
Figures


References
-
- Plato . In: The Trial and Death of Socrates. Warrington J., Translator J.M., editors. Volume 457 Dent and Sons Ltd.; London, UK: 1963. Everyman’s Library.
-
- Eigen M., McCaskill J., Schuster P. Molecular quasi-species. J. Phys. Chem. 1988;92:6881–6891. doi: 10.1021/j100335a010. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

