Optical Genome Mapping: Integrating Structural Variations for Precise Homologous Recombination Deficiency Score Calculation
- PMID: 37761823
- PMCID: PMC10530691
- DOI: 10.3390/genes14091683
Optical Genome Mapping: Integrating Structural Variations for Precise Homologous Recombination Deficiency Score Calculation
Abstract
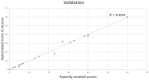
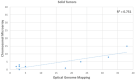
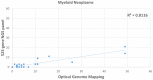
Homologous recombination deficiency (HRD) is characterized by the inability of a cell to repair the double-stranded breaks using the homologous recombination repair (HRR) pathway. The deficiency of the HRR pathway results in defective DNA repair, leading to genomic instability and tumorigenesis. The presence of HRD has been found to make tumors sensitive to ICL-inducing platinum-based therapies and poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors (PARPi). However, there are no standardized methods to measure and report HRD phenotypes. Herein, we compare optical genome mapping (OGM), chromosomal microarray (CMA), and a 523-gene NGS panel for HRD score calculations. This retrospective study included the analysis of 196 samples, of which 10 were gliomas, 176 were hematological malignancy samples, and 10 were controls. The 10 gliomas were evaluated with both CMA and OGM, and 30 hematological malignancy samples were evaluated with both the NGS panel and OGM. To verify the scores in a larger cohort, 135 cases were evaluated with the NGS panel and 71 cases with OGM. The HRD scores were calculated using a combination of three HRD signatures that included loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large-scale transitions (LST). In the ten glioma cases analyzed with OGM and CMA using the same DNA (to remove any tumor percentage bias), the HRD scores (mean ± SEM) were 13.2 (±4.2) with OGM compared to 3.7 (±1.4) with CMA. In the 30 hematological malignancy cases analyzed with OGM and the 523-gene NGS panel, the HRD scores were 7.6 (±2.2) with OGM compared to 2.6 (±0.8) with the 523-gene NGS panel. OGM detected 70.8% and 66.8% of additional variants that are considered HRD signatures in gliomas and hematological malignancies, respectively. The higher sensitivity of OGM to capture HRD signature variants might enable a more accurate and precise correlation with response to PARPi and platinum-based drugs. This study reveals HRD signatures that are cryptic to current standard of care (SOC) methods used for assessing the HRD phenotype and presents OGM as an attractive alternative with higher resolution and sensitivity to accurately assess the HRD phenotype.
Keywords: HRD scores; homologous recombination deficiency; optical genome mapping.
Conflict of interest statement
R.K. has received honoraria, and/or travel funding, and/or research support from Illumina, Asuragen, QIAGEN, Perkin Elmer Inc, Bionano Genomics, Agena, Agendia, PGDx, Thermo Fisher Scientific, Cepheid, and BMS. N.S.S. owns limited personal stocks of Bionano Genomics. A.W.C.P., D.S., A.R.H. and A.C. are salaried employees at Bionano Ge-nomics Inc. All other authors have no competing interests to disclose.
Figures





References
-
- Tempero M.A. NCCN Guidelines Updates: Pancreatic Cancer. J. Natl. Compr. Cancer Netw. 2019;17:603–605. - PubMed
-
- Gradishar W.J., Moran M.S., Abraham J., Aft R., Agnese D., Allison K.H., Blair S.L., Burstein H.J., Dang C., Elias A.D. NCCN guidelines® insights: Breast cancer, version 4.2021: Featured updates to the NCCN guidelines. J. Natl. Compr. Cancer Netw. 2021;19:484–493. doi: 10.6004/jnccn.2021.0023. - DOI - PubMed
-
- Tempero M.A., Malafa M.P., Al-Hawary M., Behrman S.W., Benson A.B., Cardin D.B., Chiorean E.G., Chung V., Czito B., Del Chiaro M. Pancreatic adenocarcinoma, version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2021;19:439–457. doi: 10.6004/jnccn.2021.0017. - DOI - PubMed
-
- Armstrong D.K., Alvarez R.D., Bakkum-Gamez J.N., Barroilhet L., Behbakht K., Berchuck A., Chen L.M., Cristea M., DeRosa M., Eisenhauer E.L. Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2021;19:191–226. doi: 10.6004/jnccn.2021.0007. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

