Analysis of the Structural Dynamics of Proteins in the Ligand-Unbound and -Bound States by Diffracted X-ray Tracking
- PMID: 37762021
- PMCID: PMC10531450
- DOI: 10.3390/ijms241813717
Analysis of the Structural Dynamics of Proteins in the Ligand-Unbound and -Bound States by Diffracted X-ray Tracking
Abstract
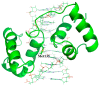
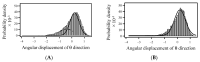
Although many protein structures have been determined at atomic resolution, the majority of them are static and represent only the most stable or averaged structures in solution. When a protein binds to its ligand, it usually undergoes fluctuation and changes its conformation. One attractive method for obtaining an accurate view of proteins in solution, which is required for applications such as the rational design of proteins and structure-based drug design, is diffracted X-ray tracking (DXT). DXT can detect the protein structural dynamics on a timeline via gold nanocrystals attached to the protein. Here, the structure dynamics of single-chain Fv antibodies, helix bundle-forming de novo designed proteins, and DNA-binding proteins in both ligand-unbound and ligand-bound states were analyzed using the DXT method. The resultant mean square angular displacements (MSD) curves in both the tilting and twisting directions clearly demonstrated that structural fluctuations were suppressed upon ligand binding, and the binding energies determined using the angular diffusion coefficients from the MSD agreed well with the binding thermodynamics determined using isothermal titration calorimetry. In addition, the size of gold nanocrystals is discussed, which is one of the technical concerns of DXT.
Keywords: DNA-binding protein; antibody; binding energy; fluctuation; helix-bundle protein.
Conflict of interest statement
The author declares no conflict of interest.
Figures
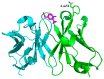

References
-
- Burley S.K., Berman H.M., Chiu W., Dai W., Flatt J.W., Hudson B.P., Kaelber J.T., Khare S.D., Kulczyk A.W., Lawson C.L., et al. Electron microscopy holdings of the Protein Data Bank: The impact of the resolution revolution, new validation tools, and implications for the future. Biophys. Rev. 2022;14:1281–1301. doi: 10.1007/s12551-022-01013-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

