Genome-Wide DNA Methylation and Gene Expression in Patients with Indolent Systemic Mastocytosis
- PMID: 37762215
- PMCID: PMC10530743
- DOI: 10.3390/ijms241813910
Genome-Wide DNA Methylation and Gene Expression in Patients with Indolent Systemic Mastocytosis
Abstract
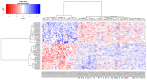
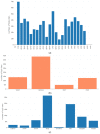
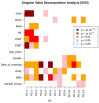
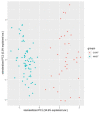
Mastocytosis is a clinically heterogenous, usually acquired disease of the mast cells with a survival time that depends on the time of onset. It ranges from skin-limited to systemic disease, including indolent and more aggressive variants. The presence of the oncogenic KIT p. D816V gene somatic mutation is a crucial element in the pathogenesis. However, further epigenetic regulation may also affect the expression of genes that are relevant to the pathology. Epigenetic alterations are responsible for regulating the expression of genes that do not modify the DNA sequence. In general, it is accepted that DNA methylation inhibits the binding of transcription factors, thereby down-regulating gene expression. However, so far, little is known about the epigenetic factors leading to the clinical onset of mastocytosis. Therefore, it is essential to identify possible epigenetic predictors, indicators of disease progression, and their link to the clinical picture to establish appropriate management and a therapeutic strategy. The aim of this study was to analyze genome-wide methylation profiles to identify differentially methylated regions (DMRs) in patients with mastocytosis compared to healthy individuals, as well as the genes located in those regulatory regions. Genome-wide DNA methylation profiling was performed in peripheral blood collected from 80 adult patients with indolent systemic mastocytosis (ISM), the most prevalent subvariant of mastocytosis, and 40 healthy adult volunteers. A total of 117 DNA samples met the criteria for the bisulfide conversion step and microarray analysis. Genome-wide DNA methylation analysis was performed using a MethylationEPIC BeadChip kit. Further analysis was focused on the genomic regions rather than individual CpG sites. Co-methylated regions (CMRs) were assigned via the CoMeBack method. To identify DMRs between the groups, a linear regression model with age as the covariate on CMRs was performed using Limma. Using the available data for cases only, an association analysis was performed between methylation status and tryptase levels, as well as the context of allergy, and anaphylaxis. KEGG pathway mapping was used to identify genes differentially expressed in anaphylaxis. Based on the DNA methylation results, the expression of 18 genes was then analyzed via real-time PCR in 20 patients with mastocytosis and 20 healthy adults. A comparison of the genome-wide DNA methylation profile between the mastocytosis patients and healthy controls revealed significant differences in the methylation levels of 85 selected CMRs. Among those, the most intriguing CMRs are 31 genes located within the regulatory regions. In addition, among the 10 CMRs located in the promoter regions, 4 and 6 regions were found to be either hypo- or hypermethylated, respectively. Importantly, three oncogenes-FOXQ1, TWIST1, and ERG-were identified as differentially methylated in mastocytosis patients, for the first time. Functional annotation revealed the most important biological processes in which the differentially methylated genes were involved as transcription, multicellular development, and signal transduction. The biological process related to histone H2A monoubiquitination (GO:0035518) was found to be enriched in association with higher tryptase levels, which may be associated with more aberrant mast cells and, therefore, more atypical mast cell disease. The signal in the BAIAP2 gene was detected in the context of anaphylaxis, but no significant differential methylation was found in the context of allergy. Furthermore, increased expression of genes encoding integral membrane components (GRM2 and KRTCAP3) was found in mastocytosis patients. This study confirms that patients with mastocytosis differ significantly in terms of methylation levels in selected CMRs of genes involved in specific molecular processes. The results of gene expression profiling indicate the increased expression of genes belonging to the integral component of the membrane in mastocytosis patients (GRM2 and KRTCAP3). Further work is warranted, especially in relation to the disease subvariants, to identify links between the methylation status and the symptoms and novel therapeutic targets.
Keywords: epigenetics; gene expression; genome-wide DNA methylation; mastocytosis.
Conflict of interest statement
The corresponding author declares no conflicts of interest regarding this publication.
Figures

Similar articles
-
DNA methylation profile in patients with indolent systemic mastocytosis.Clin Transl Allergy. 2021 Nov 2;11(9):e12074. doi: 10.1002/clt2.12074. eCollection 2021 Nov. Clin Transl Allergy. 2021. PMID: 34754417 Free PMC article.
-
Mastocytosis and related entities: a practical roadmap.Acta Clin Belg. 2023 Aug;78(4):325-335. doi: 10.1080/17843286.2022.2137631. Epub 2022 Oct 19. Acta Clin Belg. 2023. PMID: 36259506 Review.
-
KIT D816V mutation burden does not correlate to clinical manifestations of indolent systemic mastocytosis.J Allergy Clin Immunol. 2013 Sep;132(3):723-728. doi: 10.1016/j.jaci.2013.02.019. Epub 2013 Apr 12. J Allergy Clin Immunol. 2013. PMID: 23587333
-
The Number of MRGPRX2-Expressing Cells Is Increased in Skin Lesions of Patients With Indolent Systemic Mastocytosis, But Is Not Linked to Symptom Severity.Front Immunol. 2022 Jul 26;13:930945. doi: 10.3389/fimmu.2022.930945. eCollection 2022. Front Immunol. 2022. PMID: 35958589 Free PMC article.
-
Pathogenic and diagnostic relevance of KIT in primary mast cell activation disorders.Ann Allergy Asthma Immunol. 2021 Oct;127(4):427-434. doi: 10.1016/j.anai.2021.07.014. Epub 2021 Jul 20. Ann Allergy Asthma Immunol. 2021. PMID: 34298172 Review.
Cited by
-
Shake It Up Baby Now: The Changing Focus on TWIST1 and Epithelial to Mesenchymal Transition in Cancer and Other Diseases.Int J Mol Sci. 2023 Dec 16;24(24):17539. doi: 10.3390/ijms242417539. Int J Mol Sci. 2023. PMID: 38139368 Free PMC article. Review.
References
-
- Valent P., Elberink J.N.O., Gorska A., Lange M., Zanotti R., van Anrooij B., Bonifacio M., Bonadonna P., Gleixner K.V., Hadzijusufovic E., et al. The Data Registry of the European Competence Network on Mastocytosis (ECNM): Set Up, Projects, and Perspectives. J. Allergy Clin. Immunol. Pract. 2018;7:81–87. doi: 10.1016/j.jaip.2018.09.024. - DOI - PMC - PubMed
-
- Fujiwara S., Nagai H., Jimbo H., Jimbo N., Tanaka T., Inoie M., Nishigori C. Gene expression and methylation analysis in melanomas and melanocytes from the same patient: Loss of NPM2 expression is a potential immunohistochemical marker for melanoma. Front. Oncol. 2019;8:675. doi: 10.3389/fonc.2018.00675. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

