Hydroxychloroquine-Loaded Chitosan Nanoparticles Induce Anticancer Activity in A549 Lung Cancer Cells: Design, BSA Binding, Molecular Docking, Mechanistic, and Biological Evaluation
- PMID: 37762406
- PMCID: PMC10531786
- DOI: 10.3390/ijms241814103
Hydroxychloroquine-Loaded Chitosan Nanoparticles Induce Anticancer Activity in A549 Lung Cancer Cells: Design, BSA Binding, Molecular Docking, Mechanistic, and Biological Evaluation
Abstract
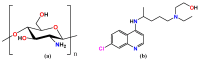
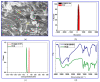
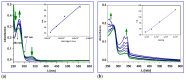
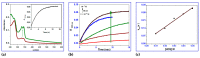
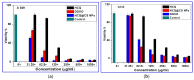
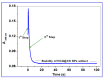
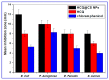
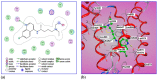
The current study describes the encapsulation of hydroxychloroquine, widely used in traditional medicine due to its diverse pharmacological and medicinal uses, in chitosan nanoparticles (CNPs). This work aims to combine the HCQ drug with CS NPs to generate a novel nanocomposite with improved characteristics and bioavailability. HCQ@CS NPs are roughly shaped like roadways and have a smooth surface with an average size of 159.3 ± 7.1 nm, a PDI of 0.224 ± 0.101, and a zeta potential of +46.6 ± 0.8 mV. To aid in the development of pharmaceutical systems for use in cancer therapy, the binding mechanism and affinity of the interaction between HCQ and HCQ@CS NPs and BSA were examined using stopped-flow and other spectroscopic approaches, supplemented by molecular docking analysis. HCQ and HCQ@CS NPs binding with BSA is driven by a ground-state complex formation that may be accompanied by a non-radiative energy transfer process, and binding constants indicate that HCQ@CS NPs-BSA was more stable than HCQ-BSA. The stopped-flow analysis demonstrated that, in addition to increasing BSA affinity, the nanoformulation HCQ@CS NPS changes the binding process and may open new routes for interaction. Docking experiments verified the development of the HCQ-BSA complex, with HCQ binding to site I on the BSA structure, primarily with the amino acids, Thr 578, Gln 579, Gln 525, Tyr 400, and Asn 404. Furthermore, the nanoformulation HCQ@CS NPS not only increased cytotoxicity against the A549 lung cancer cell line (IC50 = 28.57 ± 1.72 μg/mL) compared to HCQ (102.21 ± 0.67 μg/mL), but also exhibited higher antibacterial activity against both Gram-positive and Gram-negative bacteria when compared to HCQ and chloramphenicol, which is in agreement with the binding constants. The nanoformulation developed in this study may offer a viable therapy option for A549 lung cancer.
Keywords: A549 lung cancer; antibacterial; cell penetration; dissociation constants; drug affinity; hydroxychloroquine; molecular docking; nanoparticles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Casaluce F., Sgambato A., Maione P. The potential role of new targeted therapies in the treatment of advanced non-small-cell lung cancer. Clin. Investig. 2013;3:369–383. doi: 10.4155/cli.13.14. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

