AMMVF-DTI: A Novel Model Predicting Drug-Target Interactions Based on Attention Mechanism and Multi-View Fusion
- PMID: 37762445
- PMCID: PMC10531525
- DOI: 10.3390/ijms241814142
AMMVF-DTI: A Novel Model Predicting Drug-Target Interactions Based on Attention Mechanism and Multi-View Fusion
Abstract
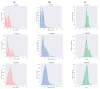
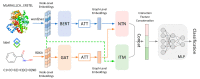
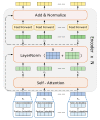
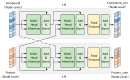
Accurate identification of potential drug-target interactions (DTIs) is a crucial task in drug development and repositioning. Despite the remarkable progress achieved in recent years, improving the performance of DTI prediction still presents significant challenges. In this study, we propose a novel end-to-end deep learning model called AMMVF-DTI (attention mechanism and multi-view fusion), which leverages a multi-head self-attention mechanism to explore varying degrees of interaction between drugs and target proteins. More importantly, AMMVF-DTI extracts interactive features between drugs and proteins from both node-level and graph-level embeddings, enabling a more effective modeling of DTIs. This advantage is generally lacking in existing DTI prediction models. Consequently, when compared to many of the start-of-the-art methods, AMMVF-DTI demonstrated excellent performance on the human, C. elegans, and DrugBank baseline datasets, which can be attributed to its ability to incorporate interactive information and mine features from both local and global structures. The results from additional ablation experiments also confirmed the importance of each module in our AMMVF-DTI model. Finally, a case study is presented utilizing our model for COVID-19-related DTI prediction. We believe the AMMVF-DTI model can not only achieve reasonable accuracy in DTI prediction, but also provide insights into the understanding of potential interactions between drugs and targets.
Keywords: drug repositioning; drug–target interaction; graph attention networks; multi-head self-attention mechanism; neural tensor networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ezzat A., Wu M., Li X., Kwoh C. Computational prediction of drug-target interactions using chemogenomic approaches: An empirical survey. Brief Bioinform. 2019;20:1337–1357. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

