Genome-Wide Identification, Characterization and Expression Analysis of the TaDUF724 Gene Family in Wheat (Triticum aestivum)
- PMID: 37762550
- PMCID: PMC10531524
- DOI: 10.3390/ijms241814248
Genome-Wide Identification, Characterization and Expression Analysis of the TaDUF724 Gene Family in Wheat (Triticum aestivum)
Abstract
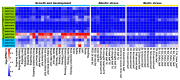
Unknown functional domain (DUF) proteins constitute a large number of functionally uncharacterized protein families in eukaryotes. DUF724s play crucial roles in plants. However, the insight understanding of wheat TaDUF724s is currently lacking. To explore the possible function of TaDUF724s in wheat growth and development and stress response, the family members were systematically identified and characterized. In total, 14 TaDUF724s were detected from a wheat reference genome; they are unevenly distributed across the 11 chromosomes, and, according to chromosome location, they were named TaDUF724-1 to TaDUF724-14. Evolution analysis revealed that TaDUF724s were under negative selection, and fragment replication was the main reason for family expansion. All TaDUF724s are unstable proteins; most TaDUF724s are acidic and hydrophilic. They were predicted to be located in the nucleus and chloroplast. The promoter regions of TaDUF724s were enriched with the cis-elements functionally associated with growth and development, as well as being hormone-responsive. Expression profiling showed that TaDUF724-9 was highly expressed in seedings, roots, leaves, stems, spikes and grains, and strongly expressed throughout the whole growth period. The 12 TaDUF724 were post-transcription regulated by 12 wheat MicroRNA (miRNA) through cleavage and translation. RT-qPCR showed that six TaDUF724s were regulated by biological and abiotic stresses. Conclusively, TaDUF724s were systematically analyzed using bioinformatics methods, which laid a theoretical foundation for clarifying the function of TaDUF724s in wheat.
Keywords: RT-qPCR; bioinformatics; cis-element; expression profiling; gene structure; miRNA; subcellular localization.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures







References
-
- Khan M.S., Rizvi A., Saif S., Zaidi A. Phosphate-solubilizing microorganisms in sustainable production of wheat: Current perspective. Probiot. Agroecosyst. 2017:51–81. doi: 10.1007/978-981-10-4059-7_3. - DOI
-
- Jiang W., Geng Y., Liu Y., Chen S., Cao S., Li W., Chen H., Ma D., Yin J. Genome-wide identification and characterization of SRO gene family in wheat: Molecular evolution and expression profiles during different stresses. Plant Physiol. Biochem. 2020;154:590–611. doi: 10.1016/j.plaphy.2020.07.006. - DOI - PubMed
-
- Yin J.L., Fang Z.W., Sun C., Zhang P., Zhang X., Lu C., Wang S.P., Ma D.F., Zhu Y.X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018;8:3086. doi: 10.1038/s41598-018-21489-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

