Chitinolytic and Fungicidal Potential of the Marine Bacterial Strains Habituating Pacific Ocean Regions
- PMID: 37764100
- PMCID: PMC10535946
- DOI: 10.3390/microorganisms11092255
Chitinolytic and Fungicidal Potential of the Marine Bacterial Strains Habituating Pacific Ocean Regions
Abstract
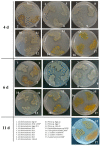
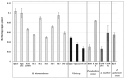
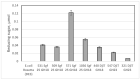
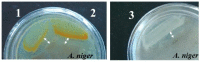
Screening for chitinolytic activity in the bacterial strains from different Pacific Ocean regions revealed that the highly active representatives belong to the genera Microbulbifer, Vibrio, Aquimarina, and Pseudoalteromonas. The widely distributed chitinolytic species was Microbulbifer isolated from the sea urchin Strongylocentrotus intermedius. Among seventeen isolates with confirmed chitinolytic activity, only the type strain P. flavipulchra KMM 3630T and the strains of putatively new species Pseudoalteromonas sp. B530 and Vibrio sp. Sgm 5, isolated from sea water (Vietnam mollusc farm) and the sea urchin S. intermedius (Peter the Great Gulf, the Sea of Japan), significantly suppressed the hyphal growth of Aspergillus niger that is perspective for the biocontrol agents' development. The results on chitinolytic activities and whole-genome sequencing of the strains under study, including agarolytic type strain Z. galactanivorans DjiT, found the new functionally active chitinase structures and biotechnological potential.
Keywords: antifungal activity; chitin-degrading enzymes; chitinase activity; functional genomics; marine chitinolytic bacteria; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Novel resources of chitinolytic bacteria isolated from Yok Don National Park, Vietnam.J Appl Microbiol. 2023 Aug 1;134(8):lxad141. doi: 10.1093/jambio/lxad141. J Appl Microbiol. 2023. PMID: 37418242
-
Isolation and identification of marine chitinolytic bacteria and their potential in antifungal biocontrol.Indian J Exp Biol. 2004 Jul;42(7):715-20. Indian J Exp Biol. 2004. PMID: 15339036
-
Characterization of Pseudoalteromonas distincta-like sea-water isolates and description of Pseudoalteromonas aliena sp. nov.Int J Syst Evol Microbiol. 2004 Sep;54(Pt 5):1431-1437. doi: 10.1099/ijs.0.03053-0. Int J Syst Evol Microbiol. 2004. PMID: 15388692
-
Marine chitinolytic enzymes, a biotechnological treasure hidden in the ocean?Appl Microbiol Biotechnol. 2018 Dec;102(23):9937-9948. doi: 10.1007/s00253-018-9385-7. Epub 2018 Oct 1. Appl Microbiol Biotechnol. 2018. PMID: 30276711 Review.
-
Bacterial Chitinase System as a Model of Chitin Biodegradation.Adv Exp Med Biol. 2019;1142:131-151. doi: 10.1007/978-981-13-7318-3_7. Adv Exp Med Biol. 2019. PMID: 31102245 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

