Impact of Environmental Sub-Inhibitory Concentrations of Antibiotics, Heavy Metals, and Biocides on the Emergence of Tolerance and Effects on the Mutant Selection Window in E. coli
- PMID: 37764108
- PMCID: PMC10535725
- DOI: 10.3390/microorganisms11092265
Impact of Environmental Sub-Inhibitory Concentrations of Antibiotics, Heavy Metals, and Biocides on the Emergence of Tolerance and Effects on the Mutant Selection Window in E. coli
Abstract
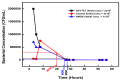
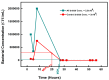
Bacteria's ability to withstand the detrimental effects of antimicrobials could occur as resistance or tolerance with the minimum inhibitory concentration, the mutant prevention concentration, and the mutant selection window as salient concepts. Thus, this study assessed the impact of exposure to extremely high doses of ampicillin on the level of persistence and tolerance development in isolates previously exposed to different concentrations of selected antibiotics, biocides, and heavy metals. These isolates were previously exposed to oxytetracycline (OXYTET), amoxicillin (AMX), copper (Cu), zinc (Zn), benzalkonium chloride (BAC) 10, dimethylammonium chloride (DADMAC) 12 and a combination of all the individual pollutants (ALL). The isolates were exposed to very high concentrations (25 × MIC) of ampicillin, and their tolerance was calculated as the time required to kill 99.9% of the bacterial population (MDK99.9). The MDK99.9 increased by 30 to 50% in test isolates (DADMAC, OXYTET, Zinc = 28 h; BAC, Copper = 30 h; amoxycillin, ALL = 26 h) compared to the untreated control. BAC-exposed isolates decreased from 2.5 × 108 CFU/mL to 2.5 × 104 CFU/mL on the second day, displaying the highest tolerance increase. The tolerance appeared to originate from two sources, i.e., stochastic persistence and genetic-induced persistence, involving multiple genes with diverse mechanisms. The mutant selection window of the isolates to ampicillin, amoxicillin, and oxytetracycline also slightly increased compared to the control, indicating the selective survival of persister cells during the 30-day exposure. These findings indicate that bacterial exposure to sub-inhibitory concentrations of environmental chemical stressors may not always result in the development of antimicrobial resistance but could initiate this process by selecting persisters that could evolve into resistant isolates.
Keywords: antibiotic resistance; environmental pollution; environmental stressors; mutation; public health; selection pressure; single nucleotide polymorphisms; tolerant bacteria.
Conflict of interest statement
Sabiha Y. Essack is the Global Respiratory Infection Partnership and a member of the Global Hygiene Council, both sponsored by unconditional educational grants from Reckitt, UK. All other authors declare that they have no conflict of interest.
Figures


Similar articles
-
Environmental concentrations of antibiotics, biocides, and heavy metals fail to induce phenotypic antimicrobial resistance in Escherichia coli.Sci Total Environ. 2023 Nov 15;899:165721. doi: 10.1016/j.scitotenv.2023.165721. Epub 2023 Jul 22. Sci Total Environ. 2023. PMID: 37482346
-
Resistance to Antibiotics, Biocides, Preservatives and Metals in Bacteria Isolated from Seafoods: Co-Selection of Strains Resistant or Tolerant to Different Classes of Compounds.Front Microbiol. 2017 Aug 31;8:1650. doi: 10.3389/fmicb.2017.01650. eCollection 2017. Front Microbiol. 2017. PMID: 28912764 Free PMC article.
-
Phenotypic zinc resistance does not correlate with antimicrobial multi-resistance in fecal E. coli isolates of piglets.Gut Pathog. 2020 Jan 21;12:4. doi: 10.1186/s13099-019-0342-5. eCollection 2020. Gut Pathog. 2020. PMID: 31988666 Free PMC article.
-
Adaptive microbial response to low-level benzalkonium chloride exposure.J Hosp Infect. 2018 Nov;100(3):e1-e22. doi: 10.1016/j.jhin.2018.05.019. Epub 2018 May 31. J Hosp Infect. 2018. PMID: 29859783 Review.
-
Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens.Antibiotics (Basel). 2015 Nov 13;4(4):567-604. doi: 10.3390/antibiotics4040567. Antibiotics (Basel). 2015. PMID: 27025641 Free PMC article. Review.
Cited by
-
Ceftazidime-avibactam tolerance and persistence among difficult-to-treat KPC-producing Klebsiella pneumoniae clinical isolates from bloodstream infections.Eur J Clin Microbiol Infect Dis. 2025 Feb;44(2):343-353. doi: 10.1007/s10096-024-05005-4. Epub 2024 Nov 30. Eur J Clin Microbiol Infect Dis. 2025. PMID: 39614972
References
-
- Van Den Bergh B., Michiels J.E., Wenseleers T., Windels E.M., Boer V., Kestemont D., De Meester L., Verstrepen K.J., Verstraeten N., Fauvart M., et al. Frequency of Antibiotic Application Drives Rapid Evolutionary Adaptation of Escherichia coli Persistence. Nat. Microbiol. 2016;1:16020. doi: 10.1038/nmicrobiol.2016.20. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

